Chapter 8 Program synthesis experiments
experiment_slug <- "2023-12-30-psynth"
working_directory <- paste0(
"experiments/",
experiment_slug,
"/analysis/"
)
# ""
if (exists("bookdown_wd_prefix")) {
working_directory <- paste0(
bookdown_wd_prefix,
working_directory
)
}8.1 Dependencies
library(tidyverse)
library(ggplot2)
library(cowplot)
library(RColorBrewer)
library(khroma)
library(rstatix)
library(knitr)
library(kableExtra)##
## Attaching package: 'kableExtra'## The following object is masked from 'package:dplyr':
##
## group_rowssource("https://gist.githubusercontent.com/benmarwick/2a1bb0133ff568cbe28d/raw/fb53bd97121f7f9ce947837ef1a4c65a73bffb3f/geom_flat_violin.R")print(version)## _
## platform aarch64-apple-darwin20
## arch aarch64
## os darwin20
## system aarch64, darwin20
## status
## major 4
## minor 2.1
## year 2022
## month 06
## day 23
## svn rev 82513
## language R
## version.string R version 4.2.1 (2022-06-23)
## nickname Funny-Looking Kid8.2 Setup
# Configure our default graphing theme
theme_set(theme_cowplot())
# Create a directory to store plots
plot_directory <- paste0(working_directory, "plots/")
dir.create(plot_directory, showWarnings=FALSE)8.2.1 Load summary data
summary_data_loc <- paste0(working_directory, "data/aggregate.csv")
summary_data <- read_csv(summary_data_loc)## Rows: 5000 Columns: 73
## ── Column specification ───────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
## Delimiter: ","
## chr (11): ANCESTOR_FILE_PATH, EVAL_FIT_EST_MODE, EVAL_MODE, POP_INIT_MODE, P...
## dbl (62): EVAL_CPU_CYCLES_PER_TEST, EVAL_MAX_PHYLO_SEARCH_DEPTH, MAX_ACTIVE_...
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.summary_data <- summary_data %>%
mutate(
eval_mode_row = case_when(
EVAL_MODE == "full" & TEST_DOWNSAMPLE_RATE == "1" ~ "down-sample",
EVAL_MODE == "full" & NUM_COHORTS == "1" ~ "cohort",
.default = EVAL_MODE
),
evals_per_gen = case_when(
EVAL_MODE == "cohort" ~ 1.0 / NUM_COHORTS,
EVAL_MODE == "down-sample" ~ TEST_DOWNSAMPLE_RATE,
EVAL_MODE == "indiv-rand-sample" ~ TEST_DOWNSAMPLE_RATE,
EVAL_MODE == "phylo-informed-sample" ~ TEST_DOWNSAMPLE_RATE,
EVAL_MODE == "full" ~ 1.0
),
EVAL_FIT_EST_MODE = case_when(
EVAL_FIT_EST_MODE == "ancestor-opt" ~ "ancestor",
EVAL_FIT_EST_MODE == "relative-opt" ~ "relative",
.default = EVAL_FIT_EST_MODE
),
est_mode_with_depth = paste(
EVAL_FIT_EST_MODE,
EVAL_MAX_PHYLO_SEARCH_DEPTH,
sep = "-"
),
eval_mode_est_mode_depth = paste(
EVAL_MODE,
EVAL_FIT_EST_MODE,
EVAL_MAX_PHYLO_SEARCH_DEPTH,
sep = "-"
),
.keep = "all"
) %>%
mutate(
eval_label = case_when(
# Clean up down-sample label
EVAL_MODE == "down-sample" & EVAL_FIT_EST_MODE != "none" ~ paste("down-sample", EVAL_FIT_EST_MODE, sep="-"),
.default = EVAL_MODE
),
) %>%
mutate(
evals_per_gen = as.factor(evals_per_gen),
est_mode_with_depth = as.factor(est_mode_with_depth),
eval_mode_est_mode_depth = as.factor(eval_mode_est_mode_depth),
EVAL_MAX_PHYLO_SEARCH_DEPTH = as.factor(EVAL_MAX_PHYLO_SEARCH_DEPTH),
PROBLEM = as.factor(PROBLEM),
SELECTION = as.factor(SELECTION),
EVAL_MODE = as.factor(EVAL_MODE),
NUM_COHORTS = as.factor(NUM_COHORTS),
TEST_DOWNSAMPLE_RATE = as.factor(TEST_DOWNSAMPLE_RATE),
EVAL_FIT_EST_MODE = factor(
EVAL_FIT_EST_MODE,
levels = c(
"none",
"ancestor",
"relative"
),
labels = c(
"None",
"Ancestor",
"Relative"
)
),
.keep = "all"
)
solution_counts <- summary_data %>%
group_by(
PROBLEM,
evals_per_gen,
eval_mode_row,
EVAL_FIT_EST_MODE,
est_mode_with_depth,
eval_mode_est_mode_depth,
EVAL_MODE,
eval_label,
EVAL_MAX_PHYLO_SEARCH_DEPTH
) %>%
summarize(
solution_count = sum(found_solution == "1"),
replicates = n(),
no_solution_count = n() - sum(found_solution == "1")
)## `summarise()` has grouped output by 'PROBLEM', 'evals_per_gen', 'eval_mode_row', 'EVAL_FIT_EST_MODE', 'est_mode_with_depth', 'eval_mode_est_mode_depth',
## 'EVAL_MODE', 'eval_label'. You can override using the `.groups` argument.# print(solution_counts, n=208)
solution_table <- kable(solution_counts) %>%
kable_styling(latex_options = "striped", font_size = 25)
save_kable(solution_table, paste0(plot_directory, "solution_counts_table.pdf"))## Note that HTML color may not be displayed on PDF properly.solution_table| PROBLEM | evals_per_gen | eval_mode_row | EVAL_FIT_EST_MODE | est_mode_with_depth | eval_mode_est_mode_depth | EVAL_MODE | eval_label | EVAL_MAX_PHYLO_SEARCH_DEPTH | solution_count | replicates | no_solution_count |
|---|---|---|---|---|---|---|---|---|---|---|---|
| bouncing-balls | 0.01 | down-sample | None | none-1 | down-sample-none-1 | down-sample | down-sample | 1 | 1 | 50 | 49 |
| bouncing-balls | 0.01 | down-sample | Ancestor | ancestor-8 | down-sample-ancestor-8 | down-sample | down-sample-ancestor | 8 | 8 | 50 | 42 |
| bouncing-balls | 0.01 | indiv-rand-sample | Ancestor | ancestor-8 | indiv-rand-sample-ancestor-8 | indiv-rand-sample | indiv-rand-sample | 8 | 7 | 50 | 43 |
| bouncing-balls | 0.01 | phylo-informed-sample | Ancestor | ancestor-8 | phylo-informed-sample-ancestor-8 | phylo-informed-sample | phylo-informed-sample | 8 | 4 | 50 | 46 |
| bouncing-balls | 0.1 | down-sample | None | none-1 | down-sample-none-1 | down-sample | down-sample | 1 | 0 | 50 | 50 |
| bouncing-balls | 0.1 | down-sample | Ancestor | ancestor-8 | down-sample-ancestor-8 | down-sample | down-sample-ancestor | 8 | 4 | 50 | 46 |
| bouncing-balls | 0.1 | indiv-rand-sample | Ancestor | ancestor-8 | indiv-rand-sample-ancestor-8 | indiv-rand-sample | indiv-rand-sample | 8 | 2 | 50 | 48 |
| bouncing-balls | 0.1 | phylo-informed-sample | Ancestor | ancestor-8 | phylo-informed-sample-ancestor-8 | phylo-informed-sample | phylo-informed-sample | 8 | 3 | 50 | 47 |
| bouncing-balls | 1 | full | None | none-1 | full-none-1 | full | full | 1 | 0 | 100 | 100 |
| dice-game | 0.01 | down-sample | None | none-1 | down-sample-none-1 | down-sample | down-sample | 1 | 0 | 50 | 50 |
| dice-game | 0.01 | down-sample | Ancestor | ancestor-8 | down-sample-ancestor-8 | down-sample | down-sample-ancestor | 8 | 26 | 50 | 24 |
| dice-game | 0.01 | indiv-rand-sample | Ancestor | ancestor-8 | indiv-rand-sample-ancestor-8 | indiv-rand-sample | indiv-rand-sample | 8 | 25 | 50 | 25 |
| dice-game | 0.01 | phylo-informed-sample | Ancestor | ancestor-8 | phylo-informed-sample-ancestor-8 | phylo-informed-sample | phylo-informed-sample | 8 | 31 | 50 | 19 |
| dice-game | 0.1 | down-sample | None | none-1 | down-sample-none-1 | down-sample | down-sample | 1 | 18 | 50 | 32 |
| dice-game | 0.1 | down-sample | Ancestor | ancestor-8 | down-sample-ancestor-8 | down-sample | down-sample-ancestor | 8 | 19 | 50 | 31 |
| dice-game | 0.1 | indiv-rand-sample | Ancestor | ancestor-8 | indiv-rand-sample-ancestor-8 | indiv-rand-sample | indiv-rand-sample | 8 | 14 | 50 | 36 |
| dice-game | 0.1 | phylo-informed-sample | Ancestor | ancestor-8 | phylo-informed-sample-ancestor-8 | phylo-informed-sample | phylo-informed-sample | 8 | 18 | 50 | 32 |
| dice-game | 1 | full | None | none-1 | full-none-1 | full | full | 1 | 0 | 100 | 100 |
| fizz-buzz | 0.01 | down-sample | None | none-1 | down-sample-none-1 | down-sample | down-sample | 1 | 5 | 50 | 45 |
| fizz-buzz | 0.01 | down-sample | Ancestor | ancestor-8 | down-sample-ancestor-8 | down-sample | down-sample-ancestor | 8 | 10 | 50 | 40 |
| fizz-buzz | 0.01 | indiv-rand-sample | Ancestor | ancestor-8 | indiv-rand-sample-ancestor-8 | indiv-rand-sample | indiv-rand-sample | 8 | 26 | 50 | 24 |
| fizz-buzz | 0.01 | phylo-informed-sample | Ancestor | ancestor-8 | phylo-informed-sample-ancestor-8 | phylo-informed-sample | phylo-informed-sample | 8 | 30 | 50 | 20 |
| fizz-buzz | 0.1 | down-sample | None | none-1 | down-sample-none-1 | down-sample | down-sample | 1 | 39 | 50 | 11 |
| fizz-buzz | 0.1 | down-sample | Ancestor | ancestor-8 | down-sample-ancestor-8 | down-sample | down-sample-ancestor | 8 | 41 | 50 | 9 |
| fizz-buzz | 0.1 | indiv-rand-sample | Ancestor | ancestor-8 | indiv-rand-sample-ancestor-8 | indiv-rand-sample | indiv-rand-sample | 8 | 19 | 50 | 31 |
| fizz-buzz | 0.1 | phylo-informed-sample | Ancestor | ancestor-8 | phylo-informed-sample-ancestor-8 | phylo-informed-sample | phylo-informed-sample | 8 | 24 | 50 | 26 |
| fizz-buzz | 1 | full | None | none-1 | full-none-1 | full | full | 1 | 9 | 100 | 91 |
| for-loop-index | 0.01 | down-sample | None | none-1 | down-sample-none-1 | down-sample | down-sample | 1 | 6 | 50 | 44 |
| for-loop-index | 0.01 | down-sample | Ancestor | ancestor-8 | down-sample-ancestor-8 | down-sample | down-sample-ancestor | 8 | 44 | 50 | 6 |
| for-loop-index | 0.01 | indiv-rand-sample | Ancestor | ancestor-8 | indiv-rand-sample-ancestor-8 | indiv-rand-sample | indiv-rand-sample | 8 | 49 | 50 | 1 |
| for-loop-index | 0.01 | phylo-informed-sample | Ancestor | ancestor-8 | phylo-informed-sample-ancestor-8 | phylo-informed-sample | phylo-informed-sample | 8 | 50 | 50 | 0 |
| for-loop-index | 0.1 | down-sample | None | none-1 | down-sample-none-1 | down-sample | down-sample | 1 | 29 | 50 | 21 |
| for-loop-index | 0.1 | down-sample | Ancestor | ancestor-8 | down-sample-ancestor-8 | down-sample | down-sample-ancestor | 8 | 35 | 50 | 15 |
| for-loop-index | 0.1 | indiv-rand-sample | Ancestor | ancestor-8 | indiv-rand-sample-ancestor-8 | indiv-rand-sample | indiv-rand-sample | 8 | 32 | 50 | 18 |
| for-loop-index | 0.1 | phylo-informed-sample | Ancestor | ancestor-8 | phylo-informed-sample-ancestor-8 | phylo-informed-sample | phylo-informed-sample | 8 | 27 | 50 | 23 |
| for-loop-index | 1 | full | None | none-1 | full-none-1 | full | full | 1 | 24 | 100 | 76 |
| gcd | 0.01 | down-sample | None | none-1 | down-sample-none-1 | down-sample | down-sample | 1 | 0 | 50 | 50 |
| gcd | 0.01 | down-sample | Ancestor | ancestor-8 | down-sample-ancestor-8 | down-sample | down-sample-ancestor | 8 | 12 | 50 | 38 |
| gcd | 0.01 | indiv-rand-sample | Ancestor | ancestor-8 | indiv-rand-sample-ancestor-8 | indiv-rand-sample | indiv-rand-sample | 8 | 18 | 50 | 32 |
| gcd | 0.01 | phylo-informed-sample | Ancestor | ancestor-8 | phylo-informed-sample-ancestor-8 | phylo-informed-sample | phylo-informed-sample | 8 | 21 | 50 | 29 |
| gcd | 0.1 | down-sample | None | none-1 | down-sample-none-1 | down-sample | down-sample | 1 | 2 | 50 | 48 |
| gcd | 0.1 | down-sample | Ancestor | ancestor-8 | down-sample-ancestor-8 | down-sample | down-sample-ancestor | 8 | 11 | 50 | 39 |
| gcd | 0.1 | indiv-rand-sample | Ancestor | ancestor-8 | indiv-rand-sample-ancestor-8 | indiv-rand-sample | indiv-rand-sample | 8 | 12 | 50 | 38 |
| gcd | 0.1 | phylo-informed-sample | Ancestor | ancestor-8 | phylo-informed-sample-ancestor-8 | phylo-informed-sample | phylo-informed-sample | 8 | 12 | 50 | 38 |
| gcd | 1 | full | None | none-1 | full-none-1 | full | full | 1 | 1 | 100 | 99 |
| grade | 0.01 | down-sample | None | none-1 | down-sample-none-1 | down-sample | down-sample | 1 | 2 | 50 | 48 |
| grade | 0.01 | down-sample | Ancestor | ancestor-8 | down-sample-ancestor-8 | down-sample | down-sample-ancestor | 8 | 35 | 50 | 15 |
| grade | 0.01 | indiv-rand-sample | Ancestor | ancestor-8 | indiv-rand-sample-ancestor-8 | indiv-rand-sample | indiv-rand-sample | 8 | 40 | 50 | 10 |
| grade | 0.01 | phylo-informed-sample | Ancestor | ancestor-8 | phylo-informed-sample-ancestor-8 | phylo-informed-sample | phylo-informed-sample | 8 | 41 | 50 | 9 |
| grade | 0.1 | down-sample | None | none-1 | down-sample-none-1 | down-sample | down-sample | 1 | 46 | 50 | 4 |
| grade | 0.1 | down-sample | Ancestor | ancestor-8 | down-sample-ancestor-8 | down-sample | down-sample-ancestor | 8 | 40 | 50 | 10 |
| grade | 0.1 | indiv-rand-sample | Ancestor | ancestor-8 | indiv-rand-sample-ancestor-8 | indiv-rand-sample | indiv-rand-sample | 8 | 35 | 50 | 15 |
| grade | 0.1 | phylo-informed-sample | Ancestor | ancestor-8 | phylo-informed-sample-ancestor-8 | phylo-informed-sample | phylo-informed-sample | 8 | 29 | 50 | 21 |
| grade | 1 | full | None | none-1 | full-none-1 | full | full | 1 | 22 | 100 | 78 |
| median | 0.01 | down-sample | None | none-1 | down-sample-none-1 | down-sample | down-sample | 1 | 50 | 50 | 0 |
| median | 0.01 | down-sample | Ancestor | ancestor-8 | down-sample-ancestor-8 | down-sample | down-sample-ancestor | 8 | 40 | 50 | 10 |
| median | 0.01 | indiv-rand-sample | Ancestor | ancestor-8 | indiv-rand-sample-ancestor-8 | indiv-rand-sample | indiv-rand-sample | 8 | 45 | 50 | 5 |
| median | 0.01 | phylo-informed-sample | Ancestor | ancestor-8 | phylo-informed-sample-ancestor-8 | phylo-informed-sample | phylo-informed-sample | 8 | 47 | 50 | 3 |
| median | 0.1 | down-sample | None | none-1 | down-sample-none-1 | down-sample | down-sample | 1 | 47 | 50 | 3 |
| median | 0.1 | down-sample | Ancestor | ancestor-8 | down-sample-ancestor-8 | down-sample | down-sample-ancestor | 8 | 45 | 50 | 5 |
| median | 0.1 | indiv-rand-sample | Ancestor | ancestor-8 | indiv-rand-sample-ancestor-8 | indiv-rand-sample | indiv-rand-sample | 8 | 47 | 50 | 3 |
| median | 0.1 | phylo-informed-sample | Ancestor | ancestor-8 | phylo-informed-sample-ancestor-8 | phylo-informed-sample | phylo-informed-sample | 8 | 40 | 50 | 10 |
| median | 1 | full | None | none-1 | full-none-1 | full | full | 1 | 34 | 100 | 66 |
| small-or-large | 0.01 | down-sample | None | none-1 | down-sample-none-1 | down-sample | down-sample | 1 | 11 | 50 | 39 |
| small-or-large | 0.01 | down-sample | Ancestor | ancestor-8 | down-sample-ancestor-8 | down-sample | down-sample-ancestor | 8 | 8 | 50 | 42 |
| small-or-large | 0.01 | indiv-rand-sample | Ancestor | ancestor-8 | indiv-rand-sample-ancestor-8 | indiv-rand-sample | indiv-rand-sample | 8 | 16 | 50 | 34 |
| small-or-large | 0.01 | phylo-informed-sample | Ancestor | ancestor-8 | phylo-informed-sample-ancestor-8 | phylo-informed-sample | phylo-informed-sample | 8 | 14 | 50 | 36 |
| small-or-large | 0.1 | down-sample | None | none-1 | down-sample-none-1 | down-sample | down-sample | 1 | 28 | 50 | 22 |
| small-or-large | 0.1 | down-sample | Ancestor | ancestor-8 | down-sample-ancestor-8 | down-sample | down-sample-ancestor | 8 | 21 | 50 | 29 |
| small-or-large | 0.1 | indiv-rand-sample | Ancestor | ancestor-8 | indiv-rand-sample-ancestor-8 | indiv-rand-sample | indiv-rand-sample | 8 | 8 | 50 | 42 |
| small-or-large | 0.1 | phylo-informed-sample | Ancestor | ancestor-8 | phylo-informed-sample-ancestor-8 | phylo-informed-sample | phylo-informed-sample | 8 | 12 | 50 | 38 |
| small-or-large | 1 | full | None | none-1 | full-none-1 | full | full | 1 | 4 | 100 | 96 |
| smallest | 0.01 | down-sample | None | none-1 | down-sample-none-1 | down-sample | down-sample | 1 | 49 | 50 | 1 |
| smallest | 0.01 | down-sample | Ancestor | ancestor-8 | down-sample-ancestor-8 | down-sample | down-sample-ancestor | 8 | 47 | 50 | 3 |
| smallest | 0.01 | indiv-rand-sample | Ancestor | ancestor-8 | indiv-rand-sample-ancestor-8 | indiv-rand-sample | indiv-rand-sample | 8 | 50 | 50 | 0 |
| smallest | 0.01 | phylo-informed-sample | Ancestor | ancestor-8 | phylo-informed-sample-ancestor-8 | phylo-informed-sample | phylo-informed-sample | 8 | 50 | 50 | 0 |
| smallest | 0.1 | down-sample | None | none-1 | down-sample-none-1 | down-sample | down-sample | 1 | 47 | 50 | 3 |
| smallest | 0.1 | down-sample | Ancestor | ancestor-8 | down-sample-ancestor-8 | down-sample | down-sample-ancestor | 8 | 47 | 50 | 3 |
| smallest | 0.1 | indiv-rand-sample | Ancestor | ancestor-8 | indiv-rand-sample-ancestor-8 | indiv-rand-sample | indiv-rand-sample | 8 | 49 | 50 | 1 |
| smallest | 0.1 | phylo-informed-sample | Ancestor | ancestor-8 | phylo-informed-sample-ancestor-8 | phylo-informed-sample | phylo-informed-sample | 8 | 49 | 50 | 1 |
| smallest | 1 | full | None | none-1 | full-none-1 | full | full | 1 | 51 | 100 | 49 |
| snow-day | 0.01 | down-sample | None | none-1 | down-sample-none-1 | down-sample | down-sample | 1 | 0 | 50 | 50 |
| snow-day | 0.01 | down-sample | Ancestor | ancestor-8 | down-sample-ancestor-8 | down-sample | down-sample-ancestor | 8 | 3 | 50 | 47 |
| snow-day | 0.01 | indiv-rand-sample | Ancestor | ancestor-8 | indiv-rand-sample-ancestor-8 | indiv-rand-sample | indiv-rand-sample | 8 | 1 | 50 | 49 |
| snow-day | 0.01 | phylo-informed-sample | Ancestor | ancestor-8 | phylo-informed-sample-ancestor-8 | phylo-informed-sample | phylo-informed-sample | 8 | 1 | 50 | 49 |
| snow-day | 0.1 | down-sample | None | none-1 | down-sample-none-1 | down-sample | down-sample | 1 | 0 | 50 | 50 |
| snow-day | 0.1 | down-sample | Ancestor | ancestor-8 | down-sample-ancestor-8 | down-sample | down-sample-ancestor | 8 | 0 | 50 | 50 |
| snow-day | 0.1 | indiv-rand-sample | Ancestor | ancestor-8 | indiv-rand-sample-ancestor-8 | indiv-rand-sample | indiv-rand-sample | 8 | 1 | 50 | 49 |
| snow-day | 0.1 | phylo-informed-sample | Ancestor | ancestor-8 | phylo-informed-sample-ancestor-8 | phylo-informed-sample | phylo-informed-sample | 8 | 4 | 50 | 46 |
| snow-day | 1 | full | None | none-1 | full-none-1 | full | full | 1 | 0 | 100 | 100 |
# Summarize avg num selected
# -- Not totally great because weird stuff happens when a solution is found (population collapses, etc)
ts_data_loc <- paste0(working_directory, "data/time_series.csv")
ts_data <- read_csv(ts_data_loc)## Rows: 99773 Columns: 24
## ── Column specification ───────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
## Delimiter: ","
## chr (6): EVAL_FIT_EST_MODE, EVAL_MODE, PROBLEM, SELECTION, TESTING_SET_PATH...
## dbl (18): EVAL_MAX_PHYLO_SEARCH_DEPTH, NUM_COHORTS, SEED, TEST_DOWNSAMPLE_RA...
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.ts_data <- ts_data %>%
mutate(
eval_mode_row = case_when(
EVAL_MODE == "full" & TEST_DOWNSAMPLE_RATE == "1" ~ "down-sample",
EVAL_MODE == "full" & NUM_COHORTS == "1" ~ "cohort",
.default = EVAL_MODE
),
evals_per_gen = case_when(
EVAL_MODE == "cohort" ~ 1.0 / NUM_COHORTS,
EVAL_MODE == "down-sample" ~ TEST_DOWNSAMPLE_RATE,
EVAL_MODE == "indiv-rand-sample" ~ TEST_DOWNSAMPLE_RATE,
EVAL_MODE == "phylo-informed-sample" ~ TEST_DOWNSAMPLE_RATE,
EVAL_MODE == "full" ~ 1.0
),
EVAL_FIT_EST_MODE = case_when(
EVAL_FIT_EST_MODE == "ancestor-opt" ~ "ancestor",
EVAL_FIT_EST_MODE == "relative-opt" ~ "relative",
.default = EVAL_FIT_EST_MODE
),
est_mode_with_depth = paste(
EVAL_FIT_EST_MODE,
EVAL_MAX_PHYLO_SEARCH_DEPTH,
sep = "-"
),
eval_mode_est_mode_depth = paste(
EVAL_MODE,
EVAL_FIT_EST_MODE,
EVAL_MAX_PHYLO_SEARCH_DEPTH,
sep = "-"
),
.keep = "all"
) %>%
mutate(
eval_label = case_when(
# Clean up down-sample label
EVAL_MODE == "down-sample" & EVAL_FIT_EST_MODE != "none" ~ paste("down-sample", EVAL_FIT_EST_MODE, sep="-"),
.default = EVAL_MODE
),
) %>%
mutate(
evals_per_gen = as.factor(evals_per_gen),
est_mode_with_depth = as.factor(est_mode_with_depth),
eval_mode_est_mode_depth = as.factor(eval_mode_est_mode_depth),
EVAL_MAX_PHYLO_SEARCH_DEPTH = as.factor(EVAL_MAX_PHYLO_SEARCH_DEPTH),
PROBLEM = as.factor(PROBLEM),
SELECTION = as.factor(SELECTION),
EVAL_MODE = as.factor(EVAL_MODE),
NUM_COHORTS = as.factor(NUM_COHORTS),
TEST_DOWNSAMPLE_RATE = as.factor(TEST_DOWNSAMPLE_RATE),
EVAL_FIT_EST_MODE = factor(
EVAL_FIT_EST_MODE,
levels = c(
"none",
"ancestor",
"relative"
),
labels = c(
"None",
"Ancestor",
"Relative"
)
),
.keep = "all"
)
ts_avgs <- ts_data %>%
group_by(
SEED,
eval_label,
evals_per_gen,
PROBLEM
) %>%
summarize(
n = n(),
avg_num_unique_selected = mean(num_unique_selected),
avg_entropy_selected_ids = mean(entropy_selected_ids)
) %>%
mutate(
eval_label = as.factor(eval_label),
evals_per_gen = as.factor(evals_per_gen),
PROBLEM = as.factor(PROBLEM)
)## `summarise()` has grouped output by 'SEED', 'eval_label', 'evals_per_gen'. You
## can override using the `.groups` argument.8.3 Problem-solving success statistics
sol_stats_data <- solution_counts %>%
filter(EVAL_MODE != "full") %>%
ungroup() %>%
unite(
"grouping",
PROBLEM,
evals_per_gen,
sep="_"
) %>%
select(
grouping, eval_label, solution_count, no_solution_count
) %>%
mutate(
grouping = as.factor(grouping)
)fisher_results <- data.frame(
comparison = character(),
group1 = character(),
group2 = character(),
n = integer(),
p = double(),
p.adj = double(),
p.adj.signif = character()
)
groupings <- levels(sol_stats_data$grouping)
for (g in groupings) {
ft_results <- sol_stats_data %>%
filter(grouping == g) %>%
select(!grouping) %>%
column_to_rownames(var = "eval_label") %>%
pairwise_fisher_test(
p.adjust.method = "holm"
) %>%
add_significance("p.adj")
ft_results <- ft_results %>%
mutate(
comparison = rep(g, nrow(ft_results)),
.keep = "all"
) %>%
relocate(comparison)
fisher_results <- rbind(
fisher_results,
ft_results
)
}
fisher_results <- as.tibble(fisher_results)## Warning: `as.tibble()` was deprecated in tibble 2.0.0.
## ℹ Please use `as_tibble()` instead.
## ℹ The signature and semantics have changed, see `?as_tibble`.
## This warning is displayed once every 8 hours.
## Call `lifecycle::last_lifecycle_warnings()` to see where this warning was generated.fisher_results <- fisher_results %>%
mutate(
comparison = as.factor(comparison),
group1 = as.factor(group1),
group2 = as.factor(group2),
) %>%
group_by(
comparison
)
fisher_table <- kbl(fisher_results) %>% kable_styling()
save_kable(fisher_table, paste0(plot_directory, "stats_table.pdf"))## Note that HTML color may not be displayed on PDF properly.fisher_table| comparison | group1 | group2 | n | p | p.adj | p.adj.signif |
|---|---|---|---|---|---|---|
| bouncing-balls_0.01 | down-sample | down-sample-ancestor | 100 | 3.09e-02 | 1.85e-01 | ns |
| bouncing-balls_0.01 | down-sample | indiv-rand-sample | 100 | 5.94e-02 | 2.97e-01 | ns |
| bouncing-balls_0.01 | down-sample | phylo-informed-sample | 100 | 3.62e-01 | 1.00e+00 | ns |
| bouncing-balls_0.01 | down-sample-ancestor | indiv-rand-sample | 100 | 1.00e+00 | 1.00e+00 | ns |
| bouncing-balls_0.01 | down-sample-ancestor | phylo-informed-sample | 100 | 3.57e-01 | 1.00e+00 | ns |
| bouncing-balls_0.01 | indiv-rand-sample | phylo-informed-sample | 100 | 5.25e-01 | 1.00e+00 | ns |
| bouncing-balls_0.1 | down-sample | down-sample-ancestor | 100 | 1.17e-01 | 7.02e-01 | ns |
| bouncing-balls_0.1 | down-sample | indiv-rand-sample | 100 | 4.95e-01 | 1.00e+00 | ns |
| bouncing-balls_0.1 | down-sample | phylo-informed-sample | 100 | 2.42e-01 | 1.00e+00 | ns |
| bouncing-balls_0.1 | down-sample-ancestor | indiv-rand-sample | 100 | 6.78e-01 | 1.00e+00 | ns |
| bouncing-balls_0.1 | down-sample-ancestor | phylo-informed-sample | 100 | 1.00e+00 | 1.00e+00 | ns |
| bouncing-balls_0.1 | indiv-rand-sample | phylo-informed-sample | 100 | 1.00e+00 | 1.00e+00 | ns |
| dice-game_0.01 | down-sample | down-sample-ancestor | 100 | 0.00e+00 | 0.00e+00 | **** |
| dice-game_0.01 | down-sample | indiv-rand-sample | 100 | 0.00e+00 | 0.00e+00 | **** |
| dice-game_0.01 | down-sample | phylo-informed-sample | 100 | 0.00e+00 | 0.00e+00 | **** |
| dice-game_0.01 | down-sample-ancestor | indiv-rand-sample | 100 | 1.00e+00 | 1.00e+00 | ns |
| dice-game_0.01 | down-sample-ancestor | phylo-informed-sample | 100 | 4.19e-01 | 9.42e-01 | ns |
| dice-game_0.01 | indiv-rand-sample | phylo-informed-sample | 100 | 3.14e-01 | 9.42e-01 | ns |
| dice-game_0.1 | down-sample | down-sample-ancestor | 100 | 1.00e+00 | 1.00e+00 | ns |
| dice-game_0.1 | down-sample | indiv-rand-sample | 100 | 5.21e-01 | 1.00e+00 | ns |
| dice-game_0.1 | down-sample | phylo-informed-sample | 100 | 1.00e+00 | 1.00e+00 | ns |
| dice-game_0.1 | down-sample-ancestor | indiv-rand-sample | 100 | 3.95e-01 | 1.00e+00 | ns |
| dice-game_0.1 | down-sample-ancestor | phylo-informed-sample | 100 | 1.00e+00 | 1.00e+00 | ns |
| dice-game_0.1 | indiv-rand-sample | phylo-informed-sample | 100 | 5.21e-01 | 1.00e+00 | ns |
| fizz-buzz_0.01 | down-sample | down-sample-ancestor | 100 | 2.62e-01 | 5.24e-01 | ns |
| fizz-buzz_0.01 | down-sample | indiv-rand-sample | 100 | 8.60e-06 | 4.28e-05 | **** |
| fizz-buzz_0.01 | down-sample | phylo-informed-sample | 100 | 2.00e-07 | 1.20e-06 | **** |
| fizz-buzz_0.01 | down-sample-ancestor | indiv-rand-sample | 100 | 1.59e-03 | 4.77e-03 | ** |
| fizz-buzz_0.01 | down-sample-ancestor | phylo-informed-sample | 100 | 8.31e-05 | 3.32e-04 | *** |
| fizz-buzz_0.01 | indiv-rand-sample | phylo-informed-sample | 100 | 5.46e-01 | 5.46e-01 | ns |
| fizz-buzz_0.1 | down-sample | down-sample-ancestor | 100 | 8.03e-01 | 8.38e-01 | ns |
| fizz-buzz_0.1 | down-sample | indiv-rand-sample | 100 | 9.55e-05 | 4.78e-04 | *** |
| fizz-buzz_0.1 | down-sample | phylo-informed-sample | 100 | 3.46e-03 | 1.04e-02 |
|
| fizz-buzz_0.1 | down-sample-ancestor | indiv-rand-sample | 100 | 1.26e-05 | 7.56e-05 | **** |
| fizz-buzz_0.1 | down-sample-ancestor | phylo-informed-sample | 100 | 6.80e-04 | 2.72e-03 | ** |
| fizz-buzz_0.1 | indiv-rand-sample | phylo-informed-sample | 100 | 4.19e-01 | 8.38e-01 | ns |
| for-loop-index_0.01 | down-sample | down-sample-ancestor | 100 | 0.00e+00 | 0.00e+00 | **** |
| for-loop-index_0.01 | down-sample | indiv-rand-sample | 100 | 0.00e+00 | 0.00e+00 | **** |
| for-loop-index_0.01 | down-sample | phylo-informed-sample | 100 | 0.00e+00 | 0.00e+00 | **** |
| for-loop-index_0.01 | down-sample-ancestor | indiv-rand-sample | 100 | 1.12e-01 | 2.24e-01 | ns |
| for-loop-index_0.01 | down-sample-ancestor | phylo-informed-sample | 100 | 2.67e-02 | 8.01e-02 | ns |
| for-loop-index_0.01 | indiv-rand-sample | phylo-informed-sample | 100 | 1.00e+00 | 1.00e+00 | ns |
| for-loop-index_0.1 | down-sample | down-sample-ancestor | 100 | 2.98e-01 | 1.00e+00 | ns |
| for-loop-index_0.1 | down-sample | indiv-rand-sample | 100 | 6.82e-01 | 1.00e+00 | ns |
| for-loop-index_0.1 | down-sample | phylo-informed-sample | 100 | 8.40e-01 | 1.00e+00 | ns |
| for-loop-index_0.1 | down-sample-ancestor | indiv-rand-sample | 100 | 6.71e-01 | 1.00e+00 | ns |
| for-loop-index_0.1 | down-sample-ancestor | phylo-informed-sample | 100 | 1.49e-01 | 8.94e-01 | ns |
| for-loop-index_0.1 | indiv-rand-sample | phylo-informed-sample | 100 | 4.16e-01 | 1.00e+00 | ns |
| gcd_0.01 | down-sample | down-sample-ancestor | 100 | 2.31e-04 | 9.24e-04 | *** |
| gcd_0.01 | down-sample | indiv-rand-sample | 100 | 1.20e-06 | 5.90e-06 | **** |
| gcd_0.01 | down-sample | phylo-informed-sample | 100 | 1.00e-07 | 4.00e-07 | **** |
| gcd_0.01 | down-sample-ancestor | indiv-rand-sample | 100 | 2.75e-01 | 5.50e-01 | ns |
| gcd_0.01 | down-sample-ancestor | phylo-informed-sample | 100 | 8.81e-02 | 2.64e-01 | ns |
| gcd_0.01 | indiv-rand-sample | phylo-informed-sample | 100 | 6.82e-01 | 6.82e-01 | ns |
| gcd_0.1 | down-sample | down-sample-ancestor | 100 | 1.47e-02 | 5.88e-02 | ns |
| gcd_0.1 | down-sample | indiv-rand-sample | 100 | 7.58e-03 | 4.55e-02 |
|
| gcd_0.1 | down-sample | phylo-informed-sample | 100 | 7.58e-03 | 4.55e-02 |
|
| gcd_0.1 | down-sample-ancestor | indiv-rand-sample | 100 | 1.00e+00 | 1.00e+00 | ns |
| gcd_0.1 | down-sample-ancestor | phylo-informed-sample | 100 | 1.00e+00 | 1.00e+00 | ns |
| gcd_0.1 | indiv-rand-sample | phylo-informed-sample | 100 | 1.00e+00 | 1.00e+00 | ns |
| grade_0.01 | down-sample | down-sample-ancestor | 100 | 0.00e+00 | 0.00e+00 | **** |
| grade_0.01 | down-sample | indiv-rand-sample | 100 | 0.00e+00 | 0.00e+00 | **** |
| grade_0.01 | down-sample | phylo-informed-sample | 100 | 0.00e+00 | 0.00e+00 | **** |
| grade_0.01 | down-sample-ancestor | indiv-rand-sample | 100 | 3.56e-01 | 7.23e-01 | ns |
| grade_0.01 | down-sample-ancestor | phylo-informed-sample | 100 | 2.41e-01 | 7.23e-01 | ns |
| grade_0.01 | indiv-rand-sample | phylo-informed-sample | 100 | 1.00e+00 | 1.00e+00 | ns |
| grade_0.1 | down-sample | down-sample-ancestor | 100 | 1.48e-01 | 4.44e-01 | ns |
| grade_0.1 | down-sample | indiv-rand-sample | 100 | 9.49e-03 | 4.74e-02 |
|
| grade_0.1 | down-sample | phylo-informed-sample | 100 | 1.43e-04 | 8.58e-04 | *** |
| grade_0.1 | down-sample-ancestor | indiv-rand-sample | 100 | 3.56e-01 | 5.96e-01 | ns |
| grade_0.1 | down-sample-ancestor | phylo-informed-sample | 100 | 2.97e-02 | 1.19e-01 | ns |
| grade_0.1 | indiv-rand-sample | phylo-informed-sample | 100 | 2.98e-01 | 5.96e-01 | ns |
| median_0.01 | down-sample | down-sample-ancestor | 100 | 1.19e-03 | 7.14e-03 | ** |
| median_0.01 | down-sample | indiv-rand-sample | 100 | 5.63e-02 | 2.82e-01 | ns |
| median_0.01 | down-sample | phylo-informed-sample | 100 | 2.42e-01 | 7.26e-01 | ns |
| median_0.01 | down-sample-ancestor | indiv-rand-sample | 100 | 2.62e-01 | 7.26e-01 | ns |
| median_0.01 | down-sample-ancestor | phylo-informed-sample | 100 | 7.13e-02 | 2.85e-01 | ns |
| median_0.01 | indiv-rand-sample | phylo-informed-sample | 100 | 7.15e-01 | 7.26e-01 | ns |
| median_0.1 | down-sample | down-sample-ancestor | 100 | 7.15e-01 | 1.00e+00 | ns |
| median_0.1 | down-sample | indiv-rand-sample | 100 | 1.00e+00 | 1.00e+00 | ns |
| median_0.1 | down-sample | phylo-informed-sample | 100 | 7.13e-02 | 4.28e-01 | ns |
| median_0.1 | down-sample-ancestor | indiv-rand-sample | 100 | 7.15e-01 | 1.00e+00 | ns |
| median_0.1 | down-sample-ancestor | phylo-informed-sample | 100 | 2.62e-01 | 1.00e+00 | ns |
| median_0.1 | indiv-rand-sample | phylo-informed-sample | 100 | 7.13e-02 | 4.28e-01 | ns |
| small-or-large_0.01 | down-sample | down-sample-ancestor | 100 | 6.11e-01 | 1.00e+00 | ns |
| small-or-large_0.01 | down-sample | indiv-rand-sample | 100 | 3.68e-01 | 1.00e+00 | ns |
| small-or-large_0.01 | down-sample | phylo-informed-sample | 100 | 6.45e-01 | 1.00e+00 | ns |
| small-or-large_0.01 | down-sample-ancestor | indiv-rand-sample | 100 | 1.00e-01 | 6.00e-01 | ns |
| small-or-large_0.01 | down-sample-ancestor | phylo-informed-sample | 100 | 2.27e-01 | 1.00e+00 | ns |
| small-or-large_0.01 | indiv-rand-sample | phylo-informed-sample | 100 | 8.28e-01 | 1.00e+00 | ns |
| small-or-large_0.1 | down-sample | down-sample-ancestor | 100 | 2.30e-01 | 4.60e-01 | ns |
| small-or-large_0.1 | down-sample | indiv-rand-sample | 100 | 5.58e-05 | 3.35e-04 | *** |
| small-or-large_0.1 | down-sample | phylo-informed-sample | 100 | 2.02e-03 | 1.01e-02 |
|
| small-or-large_0.1 | down-sample-ancestor | indiv-rand-sample | 100 | 7.58e-03 | 3.03e-02 |
|
| small-or-large_0.1 | down-sample-ancestor | phylo-informed-sample | 100 | 8.81e-02 | 2.64e-01 | ns |
| small-or-large_0.1 | indiv-rand-sample | phylo-informed-sample | 100 | 4.54e-01 | 4.60e-01 | ns |
| smallest_0.01 | down-sample | down-sample-ancestor | 100 | 6.17e-01 | 1.00e+00 | ns |
| smallest_0.01 | down-sample | indiv-rand-sample | 100 | 1.00e+00 | 1.00e+00 | ns |
| smallest_0.01 | down-sample | phylo-informed-sample | 100 | 1.00e+00 | 1.00e+00 | ns |
| smallest_0.01 | down-sample-ancestor | indiv-rand-sample | 100 | 2.42e-01 | 1.00e+00 | ns |
| smallest_0.01 | down-sample-ancestor | phylo-informed-sample | 100 | 2.42e-01 | 1.00e+00 | ns |
| smallest_0.01 | indiv-rand-sample | phylo-informed-sample | 100 | 1.00e+00 | 1.00e+00 | ns |
| smallest_0.1 | down-sample | down-sample-ancestor | 100 | 1.00e+00 | 1.00e+00 | ns |
| smallest_0.1 | down-sample | indiv-rand-sample | 100 | 6.17e-01 | 1.00e+00 | ns |
| smallest_0.1 | down-sample | phylo-informed-sample | 100 | 6.17e-01 | 1.00e+00 | ns |
| smallest_0.1 | down-sample-ancestor | indiv-rand-sample | 100 | 6.17e-01 | 1.00e+00 | ns |
| smallest_0.1 | down-sample-ancestor | phylo-informed-sample | 100 | 6.17e-01 | 1.00e+00 | ns |
| smallest_0.1 | indiv-rand-sample | phylo-informed-sample | 100 | 1.00e+00 | 1.00e+00 | ns |
| snow-day_0.01 | down-sample | down-sample-ancestor | 100 | 2.42e-01 | 1.00e+00 | ns |
| snow-day_0.01 | down-sample | indiv-rand-sample | 100 | 1.00e+00 | 1.00e+00 | ns |
| snow-day_0.01 | down-sample | phylo-informed-sample | 100 | 1.00e+00 | 1.00e+00 | ns |
| snow-day_0.01 | down-sample-ancestor | indiv-rand-sample | 100 | 6.17e-01 | 1.00e+00 | ns |
| snow-day_0.01 | down-sample-ancestor | phylo-informed-sample | 100 | 6.17e-01 | 1.00e+00 | ns |
| snow-day_0.01 | indiv-rand-sample | phylo-informed-sample | 100 | 1.00e+00 | 1.00e+00 | ns |
| snow-day_0.1 | down-sample | down-sample-ancestor | 100 | 1.00e+00 | 1.00e+00 | ns |
| snow-day_0.1 | down-sample | indiv-rand-sample | 100 | 1.00e+00 | 1.00e+00 | ns |
| snow-day_0.1 | down-sample | phylo-informed-sample | 100 | 1.17e-01 | 7.02e-01 | ns |
| snow-day_0.1 | down-sample-ancestor | indiv-rand-sample | 100 | 1.00e+00 | 1.00e+00 | ns |
| snow-day_0.1 | down-sample-ancestor | phylo-informed-sample | 100 | 1.17e-01 | 7.02e-01 | ns |
| snow-day_0.1 | indiv-rand-sample | phylo-informed-sample | 100 | 3.62e-01 | 1.00e+00 | ns |
8.4 Average number of unique candidates selected
full_avgs <- ts_data %>%
filter(eval_label == "full") %>%
group_by(PROBLEM) %>%
summarize(
n = n(),
median_num_unique_selected = median(num_unique_selected),
median_entropy_selected_ids = median(entropy_selected_ids),
avg_num_unique_selected = mean(num_unique_selected),
avg_entropy_selected_ids = mean(entropy_selected_ids)
)
build_plot_summary_data <- function(
data,
response
) {
plot <- data %>%
filter(
eval_label != "full"
) %>%
ggplot(
aes_string(
x = "eval_label",
y = response,
fill = "eval_label"
)
) +
geom_flat_violin(
position = position_nudge(x = .2, y = 0),
alpha = .8,
adjust = 1.5
) +
geom_point(
mapping = aes(color = eval_label),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_y_continuous(
# limits = c(-0.5, 100)
) +
scale_fill_bright() +
scale_color_bright() +
facet_grid(
PROBLEM ~ evals_per_gen,
# nrow=2,
labeller = label_both
) +
theme(
legend.position = "none",
axis.text.x = element_text(
angle = 30,
hjust = 1
),
panel.border = element_rect(color = "gray", size = 2)
)
return(plot)
}
plt <- build_plot_summary_data(
ts_avgs,
"avg_num_unique_selected"
)
ggsave(
filename = paste0(plot_directory, "avg_num_unique_selected.pdf"),
plot = plt
)## Saving 7 x 5 in imageplt <- build_plot_summary_data(
ts_avgs,
"avg_entropy_selected_ids"
)
ggsave(
filename = paste0(plot_directory, "avg_entropy_selected_ids.pdf"),
plot = plt
)## Saving 7 x 5 in image8.5 Phylogeny-informed Trait Estimation Distance
How is trait estimation distance distributed across sampling conditions and GP problems?
Source materials for this analysis are available here.
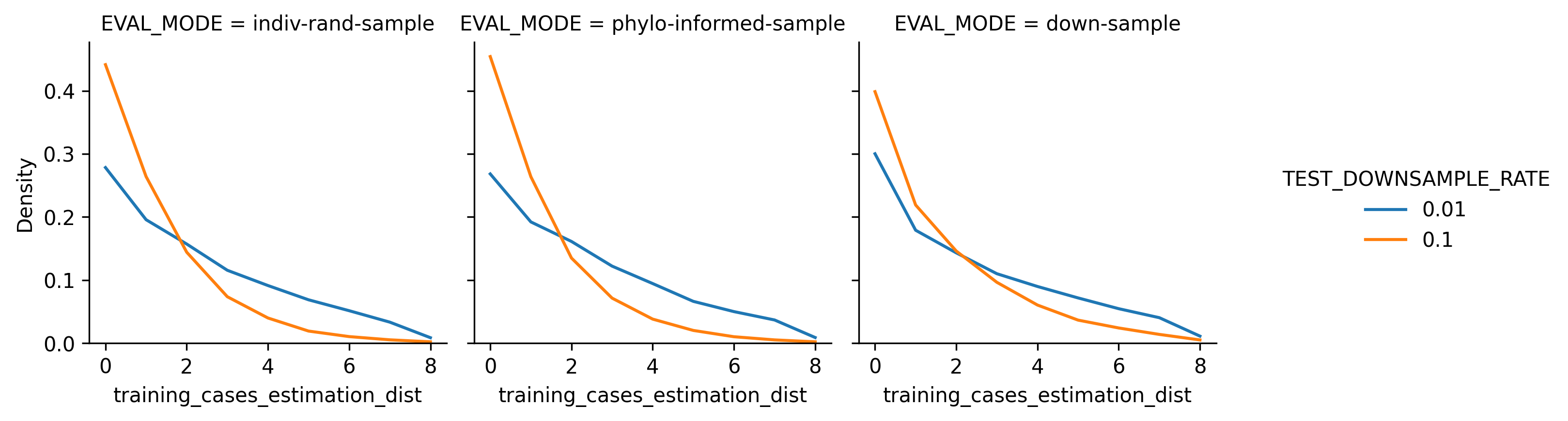
Histograms showing frequency of lookback distances for phylogeny-informed estimation.
Distribution of Estimation Distance and Downsample Rate. As would be expected, more severe downsample rates thickened the tail of longer-distance trait estmations. However, under both downsampling rates, estimations at the maximimum allowed distance of 8 generations back were rare. Note that this visualization is a histogram and does not include confidence intervals.
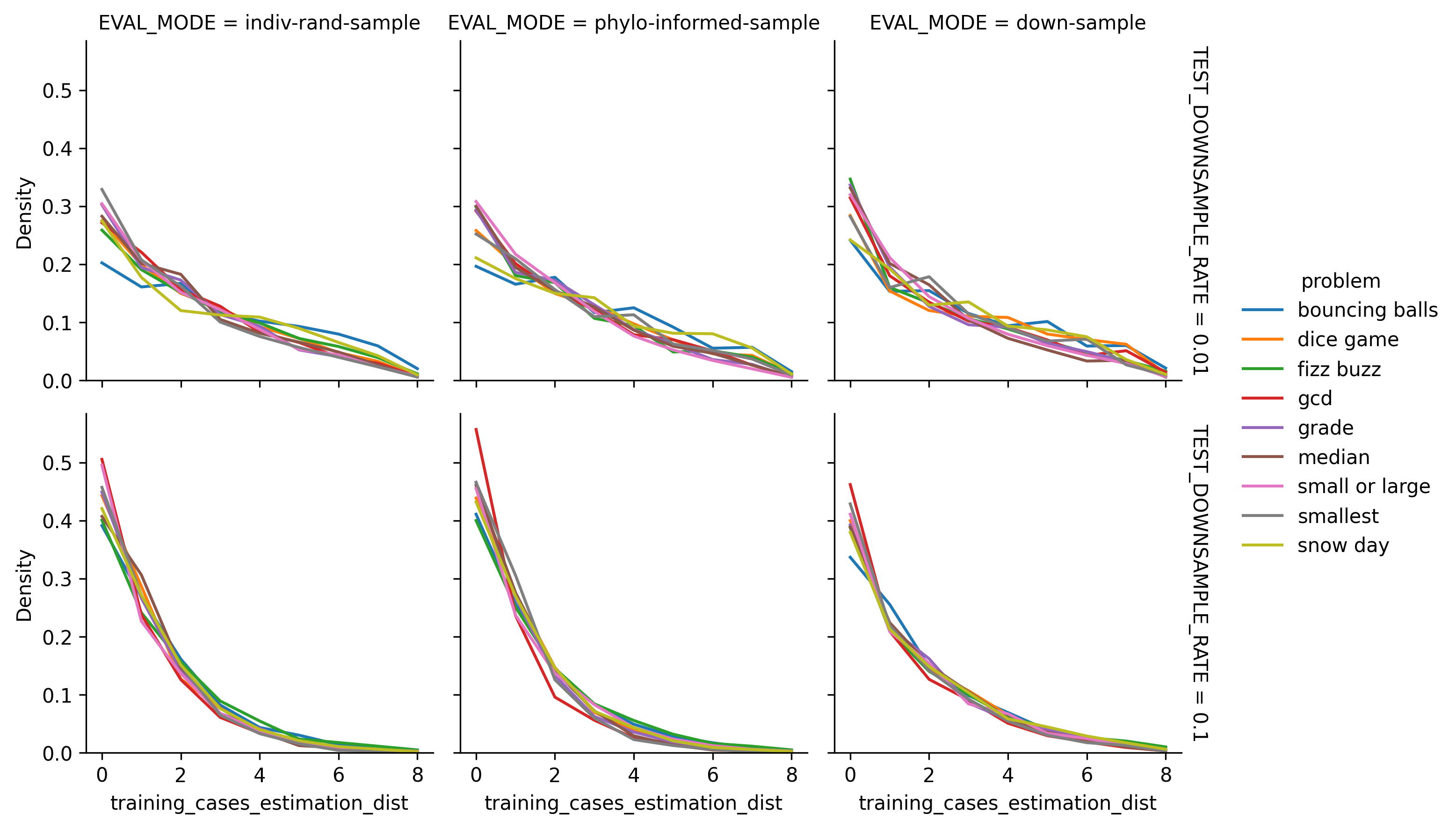
Histograms showing frequency of lookback distances for phylogeny-informed estimation.
Distribution of Estimation Distance by Program Synthesis Problem. Estimation distance distributions appear similar between problems.
8.6 Phylogeny-informed Trait Estimation Outcomes
What fraction of estimations are correct, incorrect, and failed? First visualization includes trivial (distance 0) estimations and second visualization excludes them.

Stackplots of trait estimation outcomes.
Estimation Outcomes, Including Trival Estimations. As expected, correct estimations occur less frequenetly at the severe 1% downsample rate. Except for the bouncing balls problem, more than half of estimations are correct at all downsample rates. Estimation accuracy appears to be overall roughly comparable across all downsampling methods.

Stackplots of trait estimation outcomes.
Estimation Outcomes, Excluding Trival Estimations. As expected, higher fractions of incorrect estimations are observed when including only nontrivial estimations. (Distance zero estimations are equivalent to the true trait value unless execution is nondeterministic.)
This effect is especially apparent for the bouncing balls and gcd problems under naive downsampling. At the 1% downsample rate, fewer than 25% of estimations are correct for these problems. However, as shown below, these problems are both continuous (rather than binary) with much estimation error being of small magnitude.
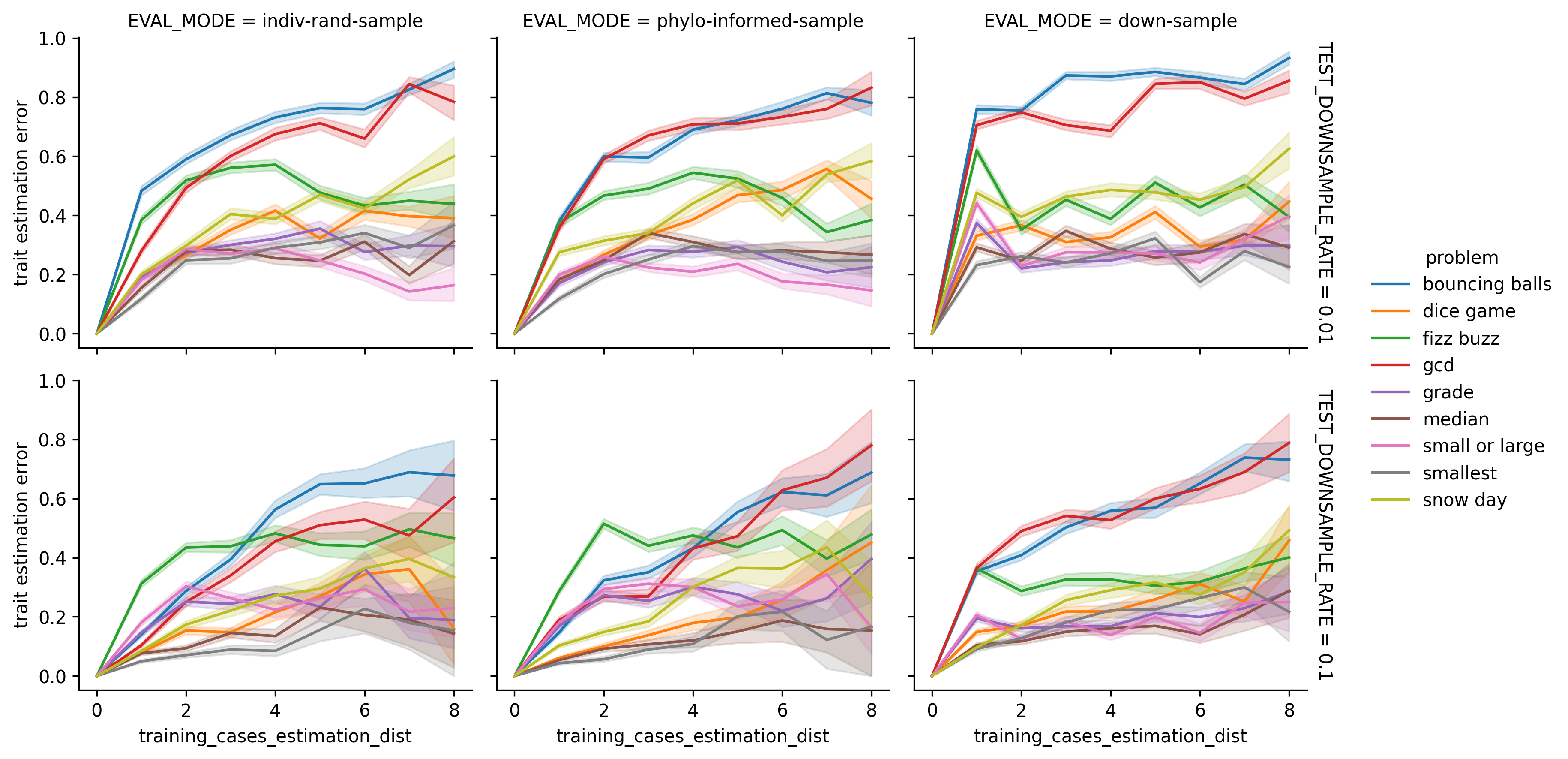
Lineplot of estimatior rates by estimation distances.
Percent Estimates Correct versus Lookup Distance. For all surveyed conditions, estimation accuracy decreases when moving from trivial distance zero estimation to nontrivial distance one estimation. However, in many (but not all) conditions the fraction of correct estimations appears to plateau past lookup distance 2. The bouncing balls and gcd problems have the highest estimation errors at long estimation distance. Interestingly, estimation error appears to spike earlier for these problems under 1% downsampling than under 10% downsampling — particularly, under naive downsampling. Other problems appear to show similar relationships between estimation error and lookup distance for both downsampling rates.
Shaded intervals are bootstrapped 95% confidence intervals. This visualization excludes failed estimations.
8.7 Distribution of Trait Estimation Error Magnidue
When trait estimation error occurs, how large is it? Note that some problems have binary traits, so the magnitude of error is limited to 0 or 1. Here, estimation error is calculated as a fraction of the true trait value.
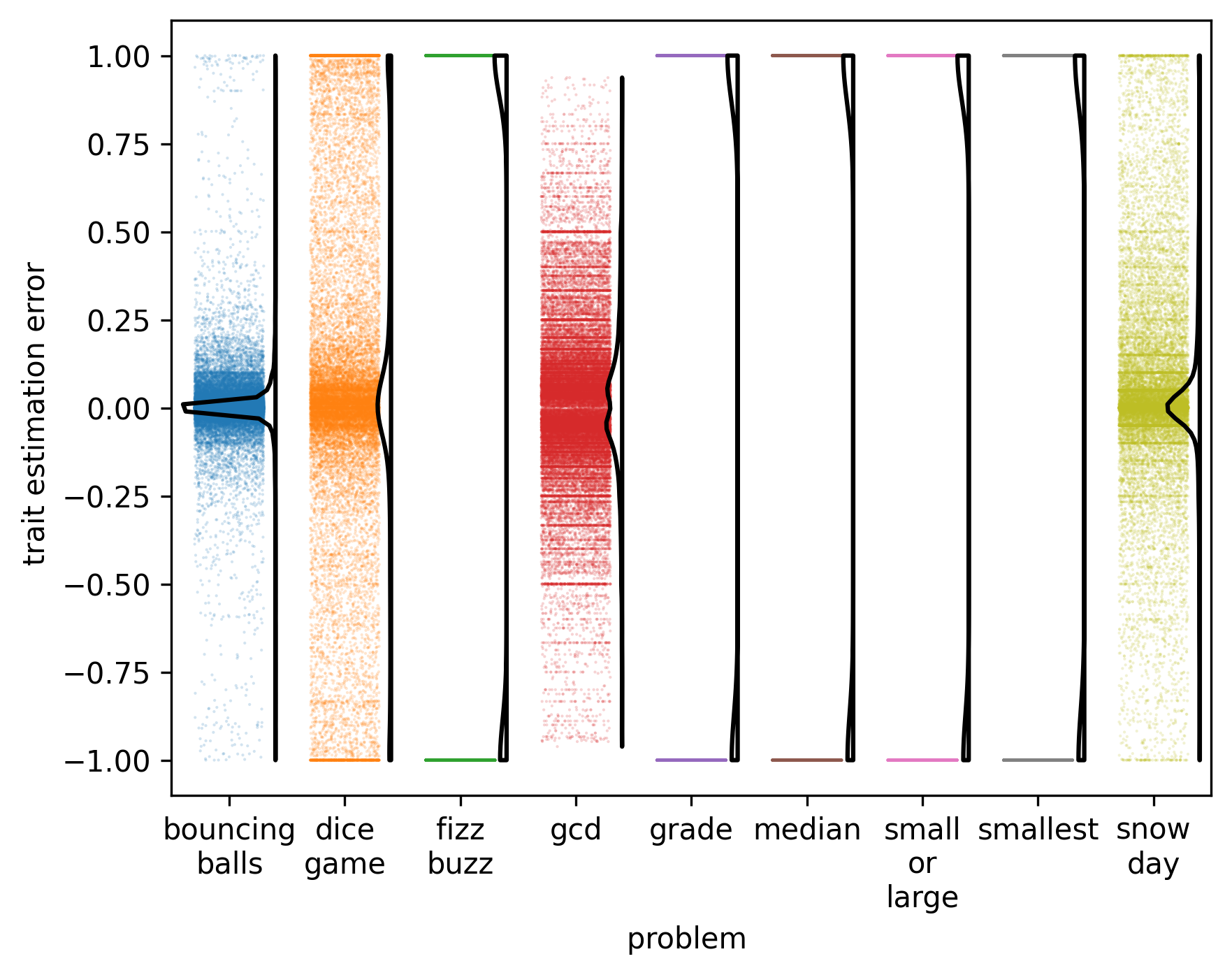
Strip plot of estimation error magnitudes.
Distribution of Estimation Error. Estimation error is generally unimodal, with bouncing balls having the tightest clustering of error near 0.0. For discrete traits, all estimation error is either -1 or 1. Note that this visualization excludes correct estimations (error magnitude exactly 0.0) and failed estimations.
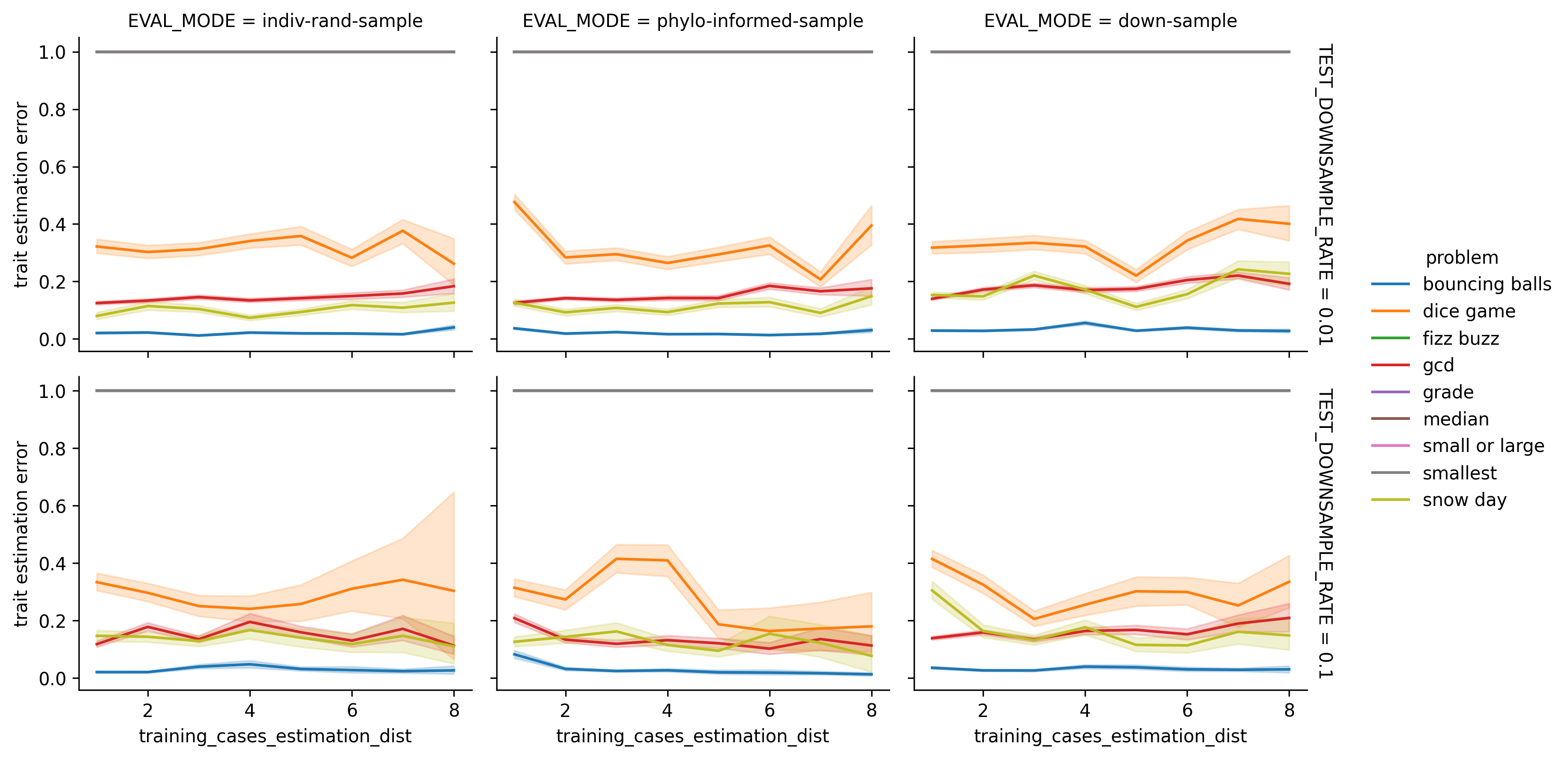
Lineplot of estimation error rates.
Trait Estimation Error versus Lookup Distance. When estimation error occurs, its magnitude is not obviously related to trait estimation distance. Note that binary traits estimation errors are all of magnitude 1.0.
Shaded intervals are bootstrapped 95% confidence intervals. This visualization excludes failed estimations.