Chapter 8 Accumulation of deleterious instructions
The effect of adaptive phenotypic plasticity on the accumulation of deleterious instructions.
8.1 Overview
total_updates <- 200000
replicates <- 100
alpha <- 0.05
focal_poison_penalty <- 0.1
focal_traits <- c("not","nand","and","ornot","or","andnot")
traits_set_a <- c("not", "and", "or")
traits_set_b <- c("nand", "ornot", "andnot")
# Relative location of data.
working_directory <- "experiments/2021-02-05-hitchhiking/analysis/" # << For bookdown
# working_directory <- "./"8.2 Analysis dependencies
Load all required R libraries.
library(RColorBrewer)
library(ggplot2)
library(rstatix)
library(ggsignif)
library(scales)
library(tidyverse)
library(cowplot)
library(Hmisc)
library(boot)
library(fmsb)
library(knitr)
source("https://gist.githubusercontent.com/benmarwick/2a1bb0133ff568cbe28d/raw/fb53bd97121f7f9ce947837ef1a4c65a73bffb3f/geom_flat_violin.R")These analyses were conducted/knitted with the following computing environment:
## _
## platform x86_64-pc-linux-gnu
## arch x86_64
## os linux-gnu
## system x86_64, linux-gnu
## status
## major 4
## minor 1.3
## year 2022
## month 03
## day 10
## svn rev 81868
## language R
## version.string R version 4.1.3 (2022-03-10)
## nickname One Push-Up8.3 Setup
####### summary data #######
summary_data_loc <- paste0(working_directory, "data/aggregate.csv")
summary_data <- read.csv(summary_data_loc, na.strings="NONE")
summary_data$DISABLE_REACTION_SENSORS <- as.factor(summary_data$DISABLE_REACTION_SENSORS)
summary_data$chg_env <- summary_data$chg_env == "True"
summary_data$dominant_plastic_odd_even <- as.factor(summary_data$dominant_plastic_odd_even)
summary_data$sensors <- summary_data$DISABLE_REACTION_SENSORS == "0"
summary_data$is_plastic <- summary_data$dominant_plastic_odd_even == "True"
summary_data$POISON_PENALTY <- as.factor(summary_data$POISON_PENALTY)
summary_data$dominant_lineage_num_times_hitchhike_inst_exec_increases_per_generation <- summary_data$dominant_lineage_num_times_hitchhike_inst_exec_increases / summary_data$dominant_generation_born
summary_data$frac_hitchhiking_linked_trait_change <- summary_data$dominant_lineage_num_times_hitchhike_inst_exec_increases_with_primary_trait_change / summary_data$dominant_lineage_num_times_hitchhike_inst_exec_increases
summary_data$frac_unexpressed_hitchhiker_inc <- summary_data$dominant_lineage_num_times_hitchhike_inst_exec_increases_in_unexpressed_phenotype / summary_data$dominant_lineage_num_times_hitchhike_inst_exec_increases
summary_data$frac_expressed_hitchiker_inc <- summary_data$dominant_lineage_num_times_hitchhike_inst_exec_increases_in_expressed_phenotype / summary_data$dominant_lineage_num_times_hitchhike_inst_exec_increases
env_label_fun <- function(chg_env) {
if (chg_env) {
return("Fluctuating")
} else {
return("Constant")
}
}
sensors_label_fun <- function(has_sensors) {
if (has_sensors) {
return("Sensors")
} else {
return("No sensors")
}
}
condition_label_fun <- function(has_sensors, env_chg) {
if (has_sensors && env_chg) {
return("PLASTIC")
} else if (env_chg) {
return("NON-PLASTIC")
} else {
return("STATIC")
}
}
summary_data$env_label <- mapply(
env_label_fun,
summary_data$chg_env
)
summary_data$sensors_label <- mapply(
sensors_label_fun,
summary_data$sensors
)
summary_data$condition <- mapply(
condition_label_fun,
summary_data$sensors,
summary_data$chg_env
)
condition_order = c(
"STATIC",
"NON-PLASTIC",
"PLASTIC"
)
pairwise_comparisons <- list(
c("STATIC", "NON-PLASTIC"),
c("STATIC", "PLASTIC"),
c("PLASTIC", "NON-PLASTIC")
)
p_label <- function(p_value) {
threshold = 0.0001
if (p_value < threshold) {
return(paste0("p < ", threshold))
} else {
return(paste0("p = ", p_value))
}
}
poison_penalties <- levels(summary_data$POISON_PENALTY)
###### time series #####
lineage_time_series_data_loc <- paste0(working_directory, "data/lineage_series.csv")
lineage_time_series_data <- read.csv(lineage_time_series_data_loc)
lineage_time_series_data$DISABLE_REACTION_SENSORS <- as.factor(lineage_time_series_data$DISABLE_REACTION_SENSORS)
lineage_time_series_data$chg_env <- lineage_time_series_data$chg_env == "True"
lineage_time_series_data$sensors <- lineage_time_series_data$DISABLE_REACTION_SENSORS == "0"
lineage_time_series_data$POISON_PENALTY <- as.factor(lineage_time_series_data$POISON_VALUE)
lineage_time_series_data$env_label <- mapply(
env_label_fun,
lineage_time_series_data$chg_env
)
lineage_time_series_data$sensors_label <- mapply(
sensors_label_fun,
lineage_time_series_data$sensors
)
lineage_time_series_data$condition <- mapply(
condition_label_fun,
lineage_time_series_data$sensors,
lineage_time_series_data$chg_env
)
####### misc #######
# Configure our default graphing theme
focal_summary_data <- filter(summary_data, POISON_PENALTY==focal_poison_penalty)
theme_set(theme_cowplot())
cb_palette <- "Paired"
dir.create(paste0(working_directory, "plots"), showWarnings=FALSE)
samplemean <- function(x, d) {
return(mean(x[d]))
}8.4 Evolution of phenotypic plasticity
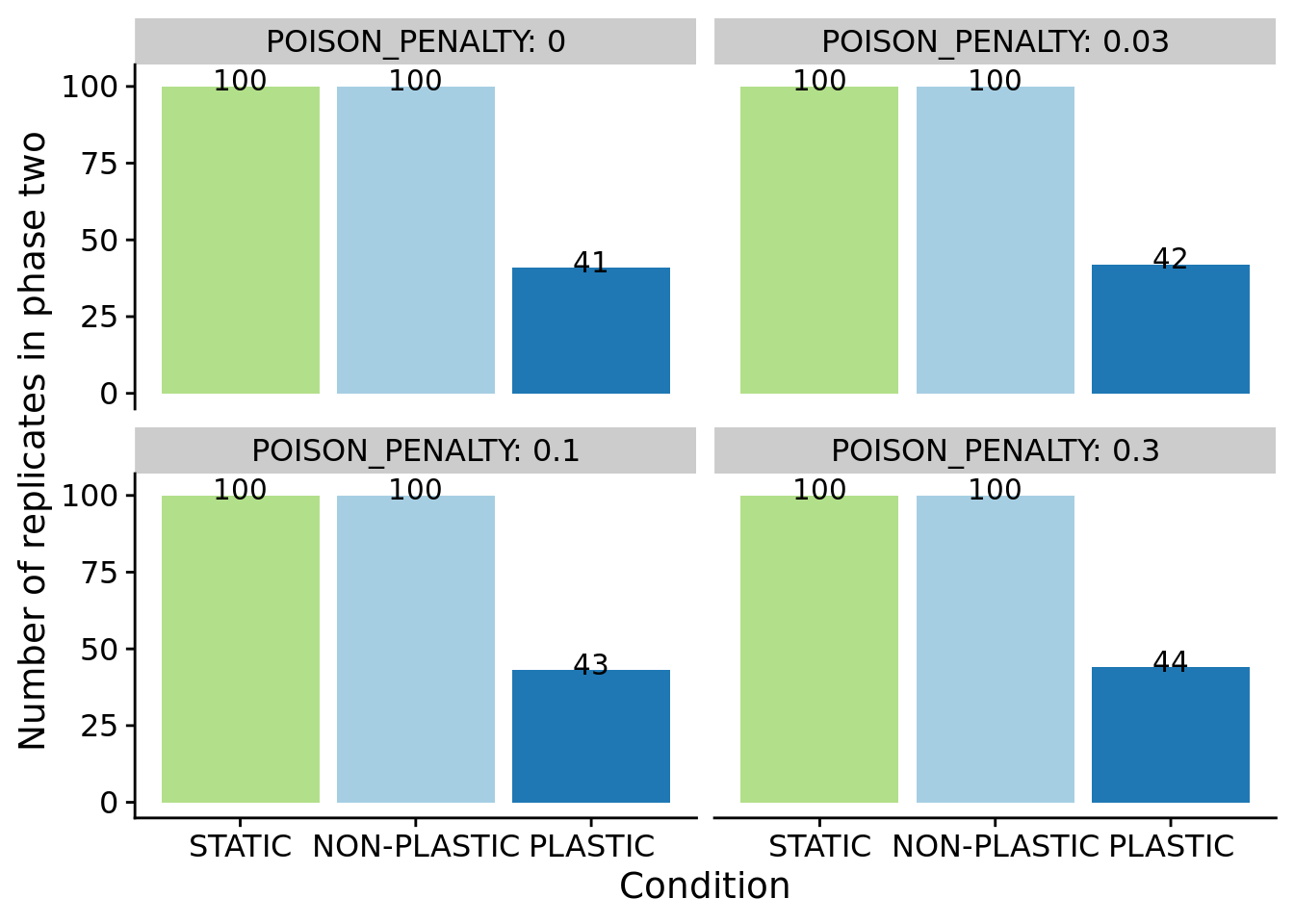
For sensor-enabled populations in fluctuating environments, we only transfered populations containing an optimally plastic genotype to phase-two.
summary_data_grouped = dplyr::group_by(summary_data, sensors, env_label, condition, POISON_PENALTY)
summary_data_group_counts = dplyr::summarize(summary_data_grouped, n=dplyr::n())
ggplot(summary_data_group_counts, aes(x=condition, y=n, fill=condition)) +
geom_col(position=position_dodge(0.9)) +
geom_text(aes(label=n, y=n+2)) +
scale_x_discrete(
name="Condition",
limits=condition_order
) +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
ylab("Number of replicates in phase two") +
facet_wrap(~POISON_PENALTY, labeller=label_both) +
theme(
legend.position="none"
)
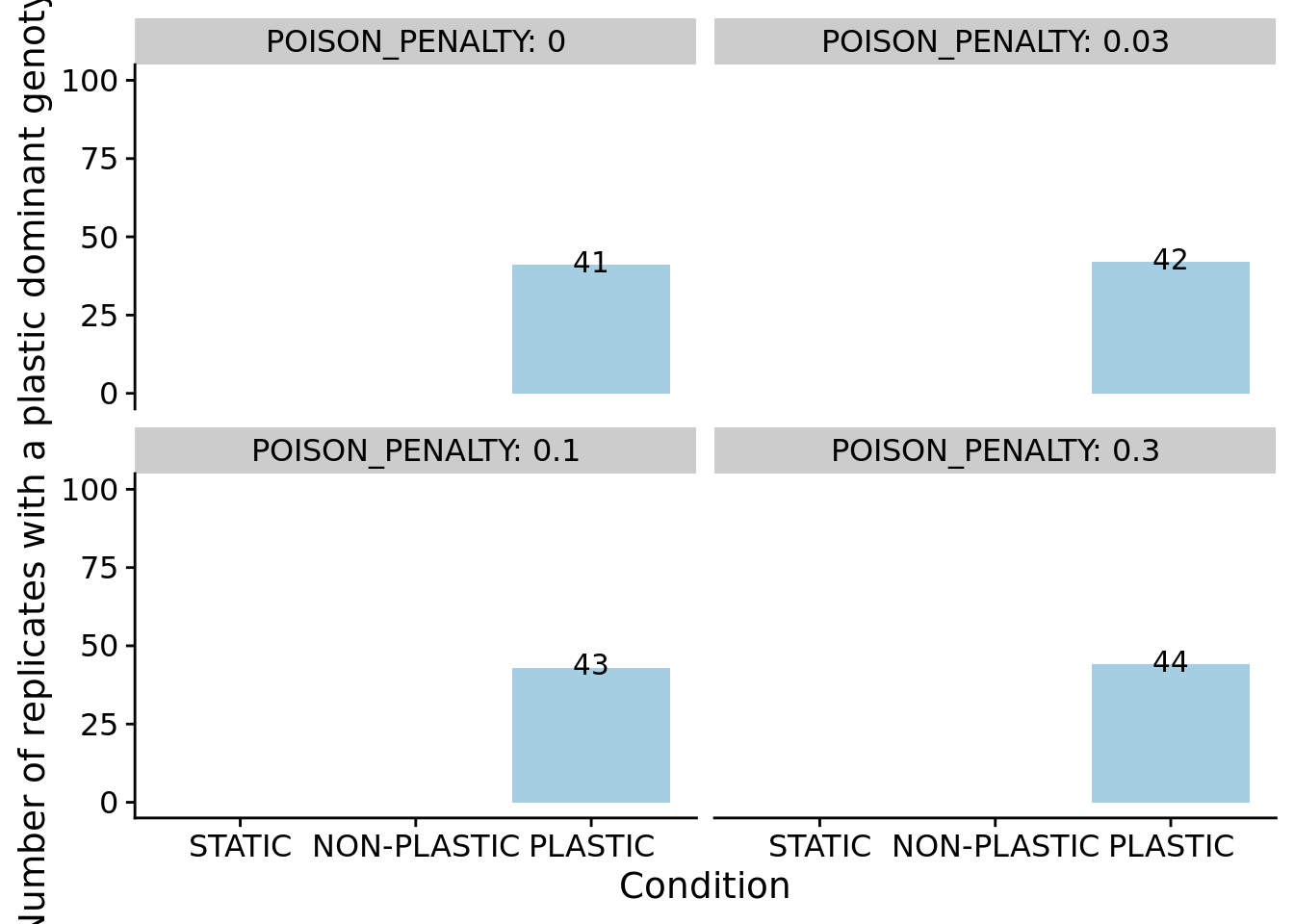
We can confirm our expectation that the dominant genotypes in non-plastic conditions are not phenotypically plastic.
summary_data_grouped = dplyr::group_by(summary_data, condition, is_plastic, POISON_PENALTY)
summary_data_group_counts = dplyr::summarize(summary_data_grouped, n=dplyr::n())## `summarise()` has grouped output by 'condition', 'is_plastic'. You can override
## using the `.groups` argument.ggplot(filter(summary_data_group_counts, is_plastic), aes(x=condition, y=n, fill=condition)) +
geom_col(position=position_dodge(0.9)) +
scale_x_discrete(
name="Condition",
limits=condition_order
) +
geom_text(aes(label=n, y=n+1)) +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
ylab("Number of replicates with a plastic dominant genotype") +
ylim(0, 100) +
facet_wrap(~POISON_PENALTY, labeller=label_both) +
theme(
legend.position="none"
)
8.5 Deleterious instruction execution
Note that under the hood, the deleterious instruction is the poison instruction in Avida.
8.5.1 Number of replicates where final dominant genotype executes the deleterious instruction
for (penalty in poison_penalties) {
occurrences <- c(
length(filter(summary_data, POISON_PENALTY==penalty & condition=="NON-PLASTIC" & dominant_times_poison_executed > 0)$RANDOM_SEED),
length(filter(summary_data, POISON_PENALTY==penalty & condition=="PLASTIC" & dominant_times_poison_executed > 0)$RANDOM_SEED),
length(filter(summary_data, POISON_PENALTY==penalty & condition=="STATIC" & dominant_times_poison_executed > 0)$RANDOM_SEED)
)
trials <- c(
length(filter(summary_data, POISON_PENALTY==penalty & condition=="NON-PLASTIC")$RANDOM_SEED),
length(filter(summary_data, POISON_PENALTY==penalty & condition=="PLASTIC")$RANDOM_SEED),
length(filter(summary_data, POISON_PENALTY==penalty & condition=="STATIC" )$RANDOM_SEED)
)
names(trials) <- c(
"NON-PLASTIC",
"PLASTIC",
"STATIC"
)
names(occurrences) <- c(
"NON-PLASTIC",
"PLASTIC",
"STATIC"
)
poison_exec_table <- data.frame(
executes.poison=occurrences,
replicates=trials
)
cat(paste0("#### Penalty: ", penalty, "\n"))
cat(print(kable(poison_exec_table)))
cat("\n")
ft <- pairwise.fisher.test(x=occurrences, n=trials, p.adjust.method="bonferroni")
print(ft)
cat("\n\n")
}## #### Penalty: 0
##
##
## | | executes.poison| replicates|
## |:-----------|---------------:|----------:|
## |NON-PLASTIC | 86| 100|
## |PLASTIC | 27| 41|
## |STATIC | 85| 100|
##
##
## Pairwise comparisons using Pairwise comparison of proportions (Fisher)
##
## data: occurrences out of trials
##
## NON-PLASTIC PLASTIC
## PLASTIC 0.03 -
## STATIC 1.00 0.06
##
## P value adjustment method: bonferroni
##
##
## #### Penalty: 0.03
##
##
## | | executes.poison| replicates|
## |:-----------|---------------:|----------:|
## |NON-PLASTIC | 46| 100|
## |PLASTIC | 1| 42|
## |STATIC | 1| 100|
##
##
## Pairwise comparisons using Pairwise comparison of proportions (Fisher)
##
## data: occurrences out of trials
##
## NON-PLASTIC PLASTIC
## PLASTIC 1.2e-07 -
## STATIC 2.9e-15 1
##
## P value adjustment method: bonferroni
##
##
## #### Penalty: 0.1
##
##
## | | executes.poison| replicates|
## |:-----------|---------------:|----------:|
## |NON-PLASTIC | 14| 100|
## |PLASTIC | 0| 43|
## |STATIC | 0| 100|
##
##
## Pairwise comparisons using Pairwise comparison of proportions (Fisher)
##
## data: occurrences out of trials
##
## NON-PLASTIC PLASTIC
## PLASTIC 0.03212 -
## STATIC 0.00022 1.00000
##
## P value adjustment method: bonferroni
##
##
## #### Penalty: 0.3
##
##
## | | executes.poison| replicates|
## |:-----------|---------------:|----------:|
## |NON-PLASTIC | 0| 100|
## |PLASTIC | 0| 44|
## |STATIC | 0| 100|
##
##
## Pairwise comparisons using Pairwise comparison of proportions (Fisher)
##
## data: occurrences out of trials
##
## NON-PLASTIC PLASTIC
## PLASTIC 1 -
## STATIC 1 1
##
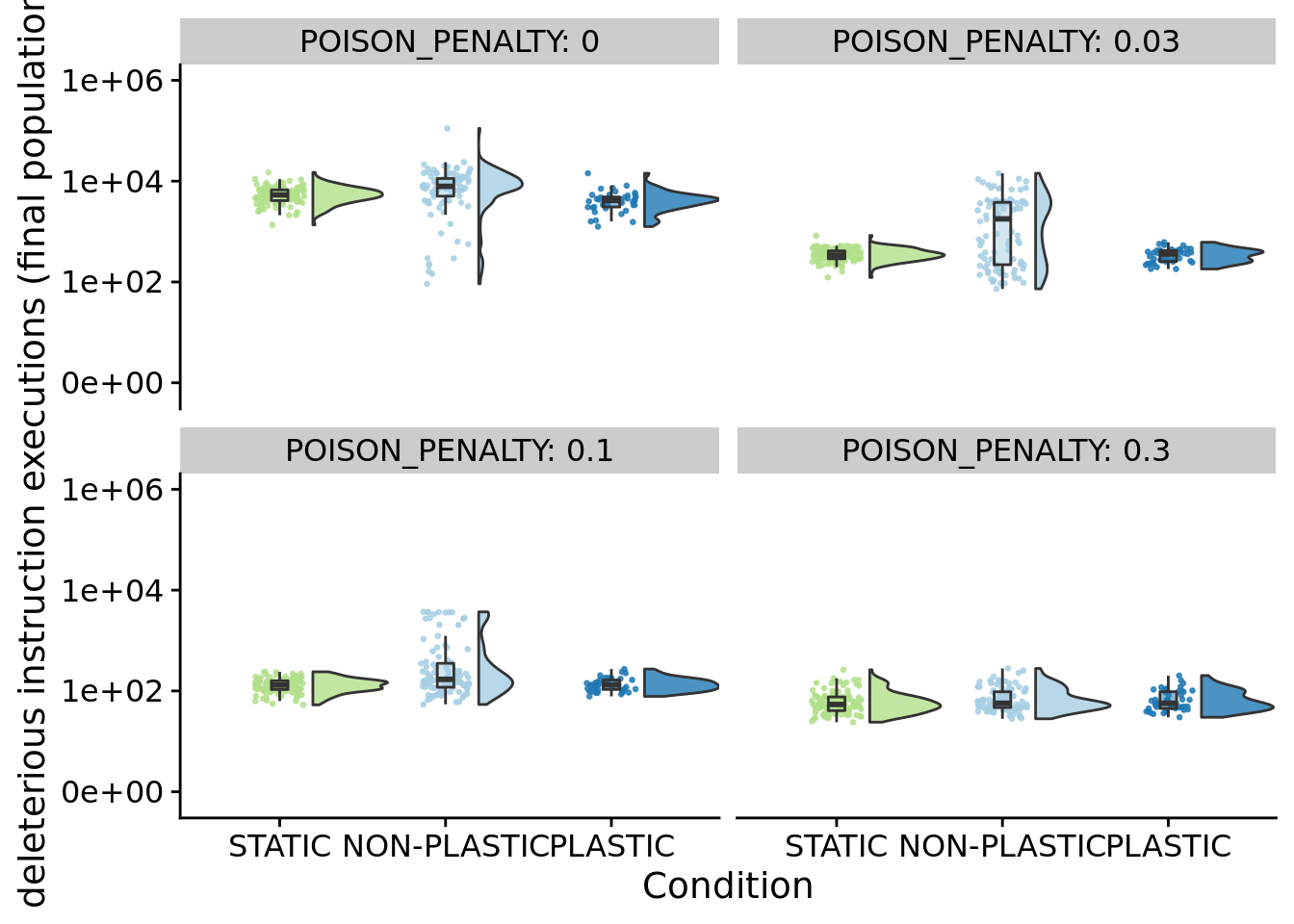
## P value adjustment method: bonferroni8.5.2 Deleterious instruction execution (final population)
ggplot(summary_data, aes(x=condition, y=final_population_poison, fill=condition)) +
geom_flat_violin(
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Condition",
limits=condition_order
) +
scale_y_continuous(
name="deleterious instruction executions (final population)",
trans=pseudo_log_trans(sigma=1,base=10),
breaks=c(0,100,10000,1000000),
limits=c(-1,1000000)
) +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
facet_wrap(
~POISON_PENALTY,
labeller=label_both
) +
# coord_flip() +
theme(
legend.position="none"
)
for (penalty in poison_penalties) {
stat_data <- filter(summary_data, POISON_PENALTY==penalty)
print(
paste0(
"PENALTY: ", penalty
)
)
kt <- kruskal.test(
formula=final_population_poison~condition,
data=stat_data
)
print(
kt
)
if (is.na(kt$p.value)) { next }
if (kt$p.value > 0.05) { next }
print(
pairwise.wilcox.test(
x=stat_data$final_population_poison,
g=stat_data$condition,
p.adjust.method="bonferroni"
)
)
}## [1] "PENALTY: 0"
##
## Kruskal-Wallis rank sum test
##
## data: final_population_poison by condition
## Kruskal-Wallis chi-squared = 43.589, df = 2, p-value = 3.426e-10
##
##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: stat_data$final_population_poison and stat_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC 8.7e-07 -
## STATIC 9.8e-07 0.00074
##
## P value adjustment method: bonferroni
## [1] "PENALTY: 0.03"
##
## Kruskal-Wallis rank sum test
##
## data: final_population_poison by condition
## Kruskal-Wallis chi-squared = 20.74, df = 2, p-value = 3.136e-05
##
##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: stat_data$final_population_poison and stat_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC 0.003 -
## STATIC 1e-04 1.000
##
## P value adjustment method: bonferroni
## [1] "PENALTY: 0.1"
##
## Kruskal-Wallis rank sum test
##
## data: final_population_poison by condition
## Kruskal-Wallis chi-squared = 20.608, df = 2, p-value = 3.35e-05
##
##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: stat_data$final_population_poison and stat_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC 0.0093 -
## STATIC 4.9e-05 1.0000
##
## P value adjustment method: bonferroni
## [1] "PENALTY: 0.3"
##
## Kruskal-Wallis rank sum test
##
## data: final_population_poison by condition
## Kruskal-Wallis chi-squared = 3.3994, df = 2, p-value = 0.18278.5.3 Cummulative deleterious instruction execution along final dominant lineages
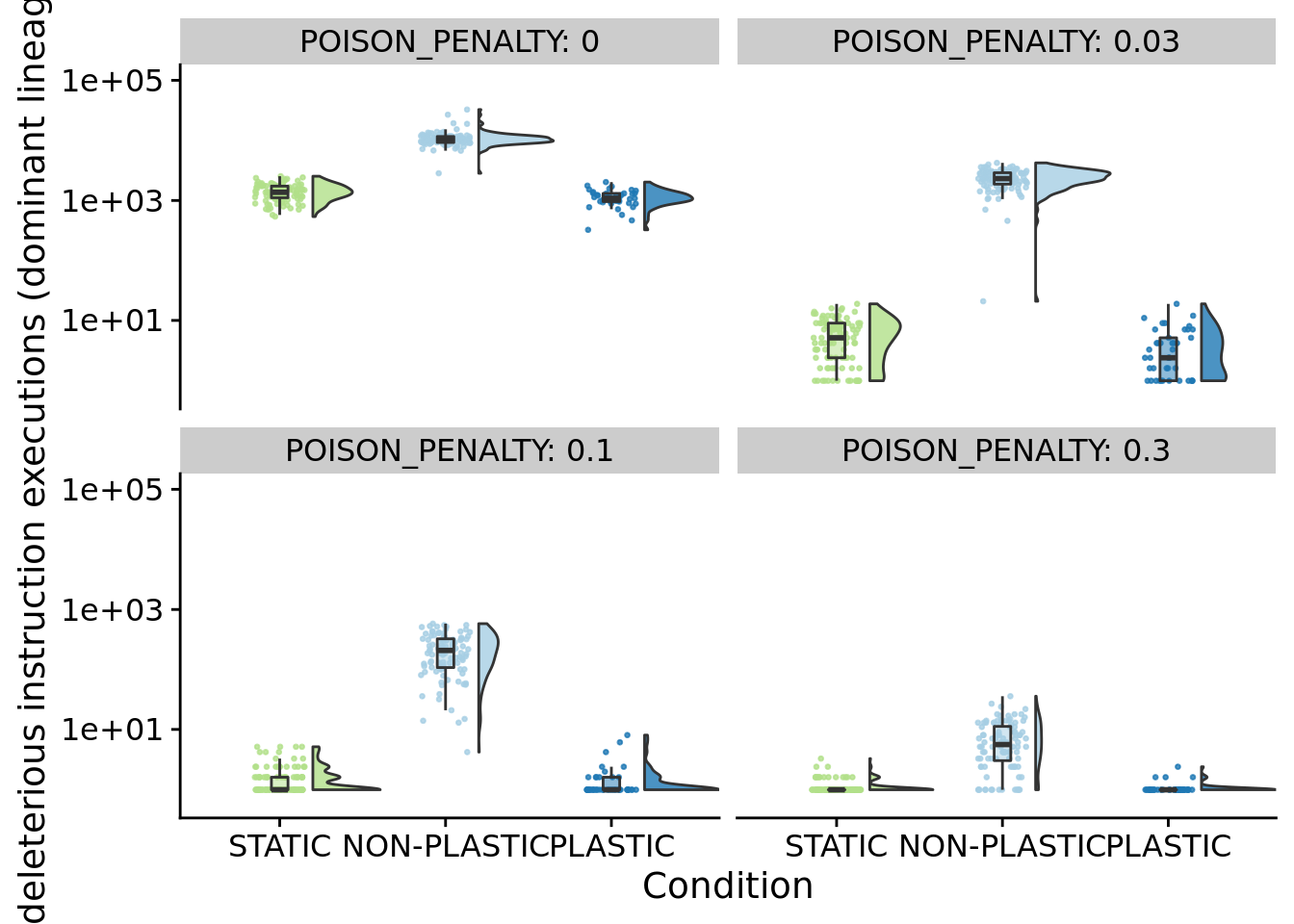
ggplot(summary_data, aes(x=condition, y=dominant_lineage_times_poison_executed, fill=condition)) +
geom_flat_violin(
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Condition",
limits=condition_order
) +
scale_y_continuous(
name="deleterious instruction executions (dominant lineage)",
trans=pseudo_log_trans(sigma = 1, base = 10),
breaks=c(10,1000,100000),
limits=c(-1,100000)
) +
facet_wrap(
~POISON_PENALTY,
labeller=label_both
) +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
theme(
legend.position="none"
)
ggsave(
paste0(working_directory, "plots/final-dominant-lineage-poison-log.pdf"),
width=15,
height=10
)for (penalty in poison_penalties) {
stat_data <- filter(summary_data, POISON_PENALTY==penalty)
print(
paste0(
"PENALTY: ", penalty
)
)
kt <- kruskal.test(
formula=dominant_lineage_times_poison_executed~condition,
data=stat_data
)
print(
kt
)
if (is.na(kt$p.value)) { next }
if (kt$p.value > 0.05) { next }
print(
pairwise.wilcox.test(
x=stat_data$dominant_lineage_times_poison_executed,
g=stat_data$condition,
p.adjust.method="bonferroni"
)
)
}## [1] "PENALTY: 0"
##
## Kruskal-Wallis rank sum test
##
## data: dominant_lineage_times_poison_executed by condition
## Kruskal-Wallis chi-squared = 178.84, df = 2, p-value < 2.2e-16
##
##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: stat_data$dominant_lineage_times_poison_executed and stat_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC <2e-16 -
## STATIC <2e-16 0.0018
##
## P value adjustment method: bonferroni
## [1] "PENALTY: 0.03"
##
## Kruskal-Wallis rank sum test
##
## data: dominant_lineage_times_poison_executed by condition
## Kruskal-Wallis chi-squared = 178.62, df = 2, p-value < 2.2e-16
##
##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: stat_data$dominant_lineage_times_poison_executed and stat_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC <2e-16 -
## STATIC <2e-16 0.011
##
## P value adjustment method: bonferroni
## [1] "PENALTY: 0.1"
##
## Kruskal-Wallis rank sum test
##
## data: dominant_lineage_times_poison_executed by condition
## Kruskal-Wallis chi-squared = 184.83, df = 2, p-value < 2.2e-16
##
##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: stat_data$dominant_lineage_times_poison_executed and stat_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC <2e-16 -
## STATIC <2e-16 0.21
##
## P value adjustment method: bonferroni
## [1] "PENALTY: 0.3"
##
## Kruskal-Wallis rank sum test
##
## data: dominant_lineage_times_poison_executed by condition
## Kruskal-Wallis chi-squared = 149.48, df = 2, p-value < 2.2e-16
##
##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: stat_data$dominant_lineage_times_poison_executed and stat_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC 4.4e-16 -
## STATIC < 2e-16 0.84
##
## P value adjustment method: bonferroni8.6 Characterizing mutations that increase deleterious instruction execution
8.6.1 Number of offspring along dominant lineage with increase in deleterious instruction execution
ggplot(summary_data, aes(x=condition, y=dominant_lineage_num_times_hitchhike_inst_exec_increases, fill=condition)) +
geom_flat_violin(
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Condition",
limits=condition_order
) +
facet_wrap(
~POISON_PENALTY,
labeller=label_both
) +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
theme(
legend.position="none"
)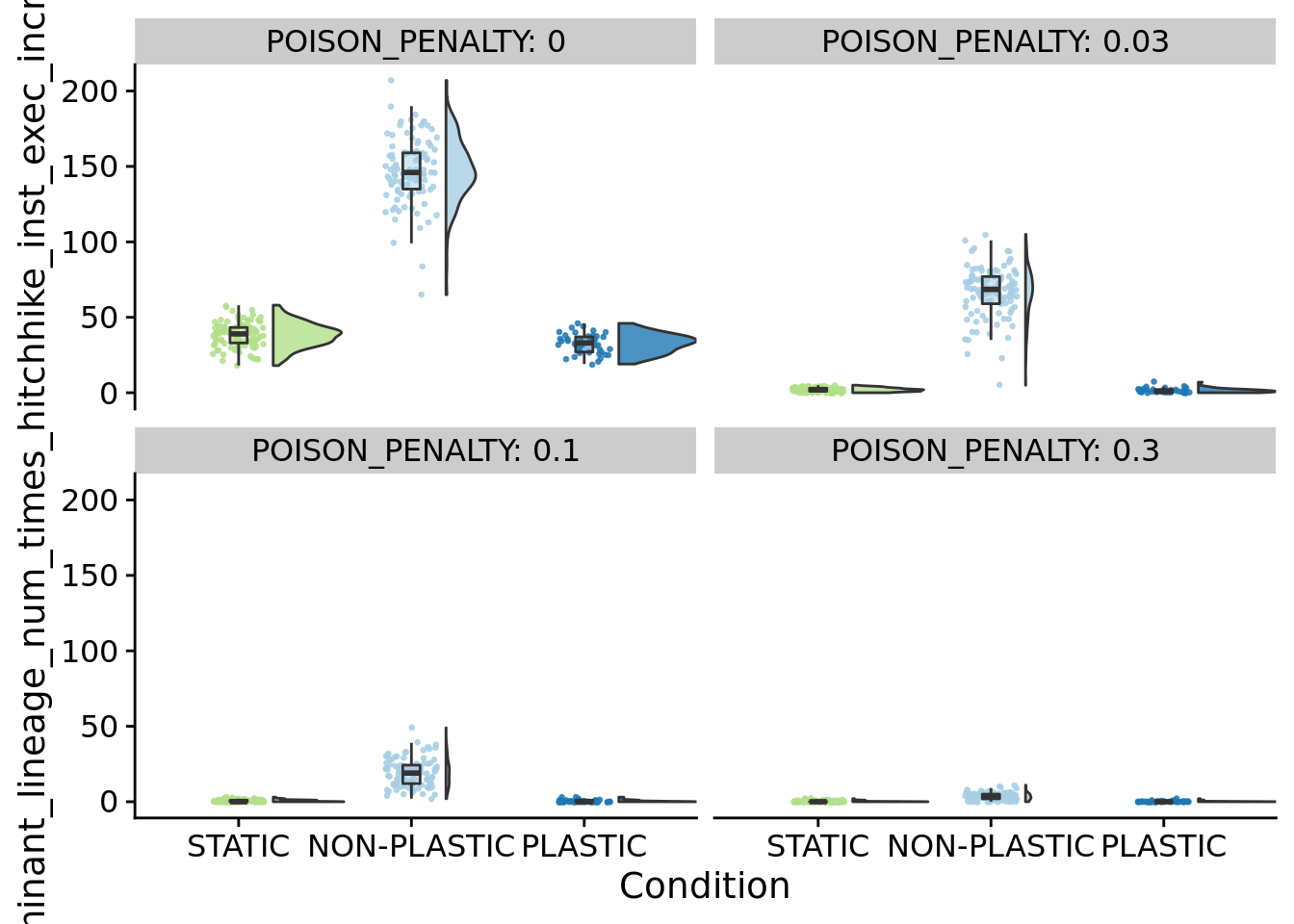
for (penalty in poison_penalties) {
stat_data <- filter(summary_data, POISON_PENALTY==penalty)
print(
paste0(
"PENALTY: ", penalty
)
)
kt <- kruskal.test(
formula=dominant_lineage_num_times_hitchhike_inst_exec_increases~condition,
data=stat_data
)
print(
kt
)
if (is.na(kt$p.value)) { next }
if (kt$p.value > 0.05) { next }
print(
pairwise.wilcox.test(
x=stat_data$dominant_lineage_num_times_hitchhike_inst_exec_increases,
g=stat_data$condition,
p.adjust.method="bonferroni"
)
)
}## [1] "PENALTY: 0"
##
## Kruskal-Wallis rank sum test
##
## data: dominant_lineage_num_times_hitchhike_inst_exec_increases by condition
## Kruskal-Wallis chi-squared = 179.79, df = 2, p-value < 2.2e-16
##
##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: stat_data$dominant_lineage_num_times_hitchhike_inst_exec_increases and stat_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC < 2e-16 -
## STATIC < 2e-16 0.00046
##
## P value adjustment method: bonferroni
## [1] "PENALTY: 0.03"
##
## Kruskal-Wallis rank sum test
##
## data: dominant_lineage_num_times_hitchhike_inst_exec_increases by condition
## Kruskal-Wallis chi-squared = 179.35, df = 2, p-value < 2.2e-16
##
##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: stat_data$dominant_lineage_num_times_hitchhike_inst_exec_increases and stat_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC <2e-16 -
## STATIC <2e-16 0.03
##
## P value adjustment method: bonferroni
## [1] "PENALTY: 0.1"
##
## Kruskal-Wallis rank sum test
##
## data: dominant_lineage_num_times_hitchhike_inst_exec_increases by condition
## Kruskal-Wallis chi-squared = 185.34, df = 2, p-value < 2.2e-16
##
##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: stat_data$dominant_lineage_num_times_hitchhike_inst_exec_increases and stat_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC <2e-16 -
## STATIC <2e-16 0.27
##
## P value adjustment method: bonferroni
## [1] "PENALTY: 0.3"
##
## Kruskal-Wallis rank sum test
##
## data: dominant_lineage_num_times_hitchhike_inst_exec_increases by condition
## Kruskal-Wallis chi-squared = 146.35, df = 2, p-value < 2.2e-16
##
##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: stat_data$dominant_lineage_num_times_hitchhike_inst_exec_increases and stat_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC 7.8e-16 -
## STATIC < 2e-16 0.86
##
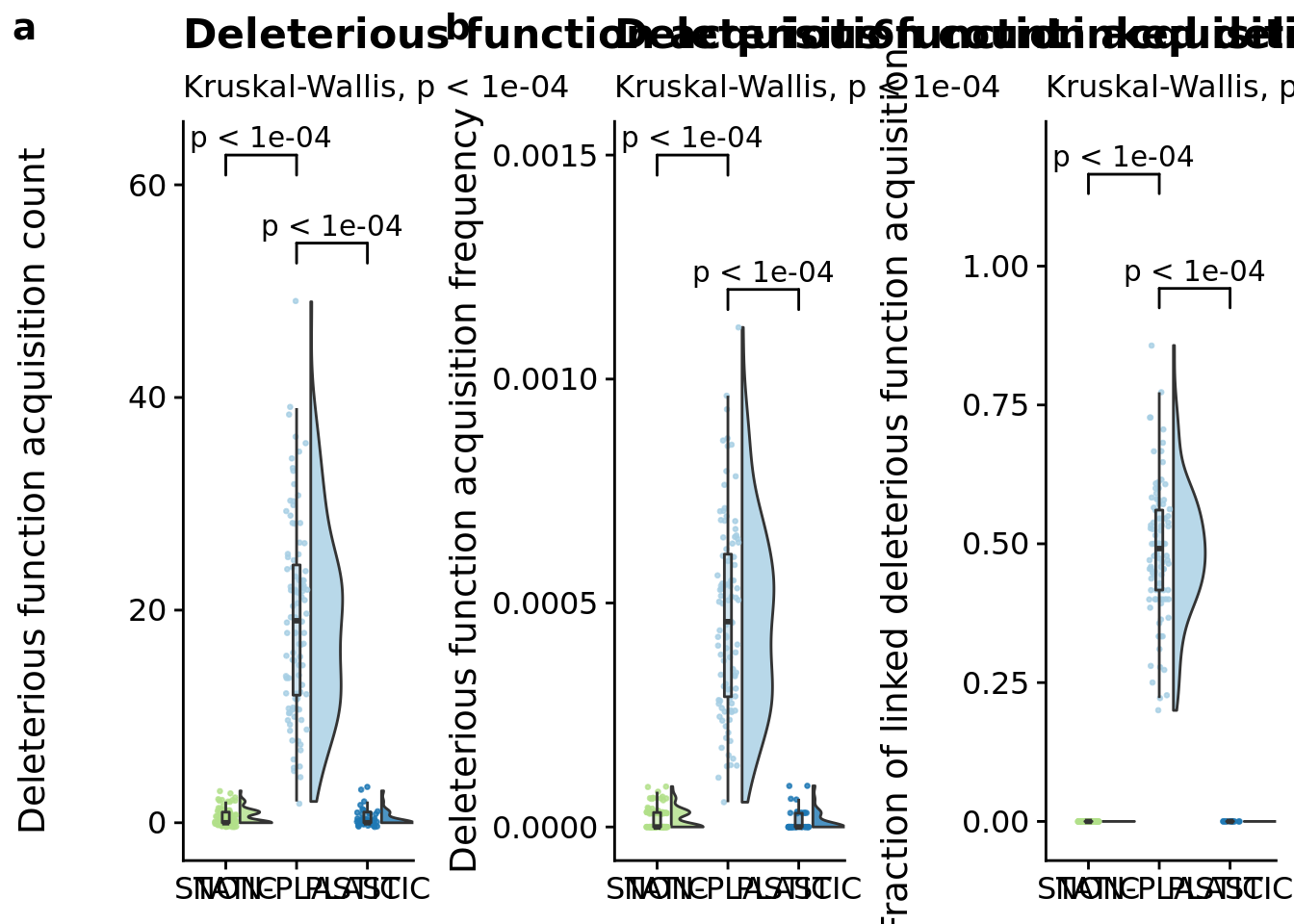
## P value adjustment method: bonferroniFocal figure for the manuscript:
# Compute manual labels for geom_signif
stat.test <- focal_summary_data %>%
wilcox_test(dominant_lineage_num_times_hitchhike_inst_exec_increases ~ condition) %>%
adjust_pvalue(method = "bonferroni") %>%
add_significance() %>%
add_xy_position(x="condition")
# Tweak y.position manually to account for scaled axis (edge case that triggers bad behavior in geom_signif)
stat.test$manual_position <- stat.test$y.position
stat.test$label <- mapply(p_label,stat.test$p.adj)
poison_increases_fig <- ggplot(
focal_summary_data,
aes(x=condition, y=dominant_lineage_num_times_hitchhike_inst_exec_increases, fill=condition)
) +
geom_flat_violin(
scale="width",
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Condition",
limits=condition_order,
labels=condition_order
) +
scale_y_continuous(
name="Deleterious function acquisition count",
) +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
# coord_flip()
labs(
subtitle=paste0(
"Kruskal-Wallis, ",
p_label(signif(kruskal.test(formula=dominant_lineage_num_times_hitchhike_inst_exec_increases~condition, data=focal_summary_data)$p.value,digits=4))
)
) +
ggsignif::geom_signif(
data=filter(stat.test, p.adj <= alpha),
aes(xmin=group1,xmax=group2,annotations=label,y_position=manual_position),
manual=TRUE,
inherit.aes=FALSE
) +
theme(
legend.position="none"
)## Warning: Ignoring unknown aesthetics: xmin, xmax, annotations, y_position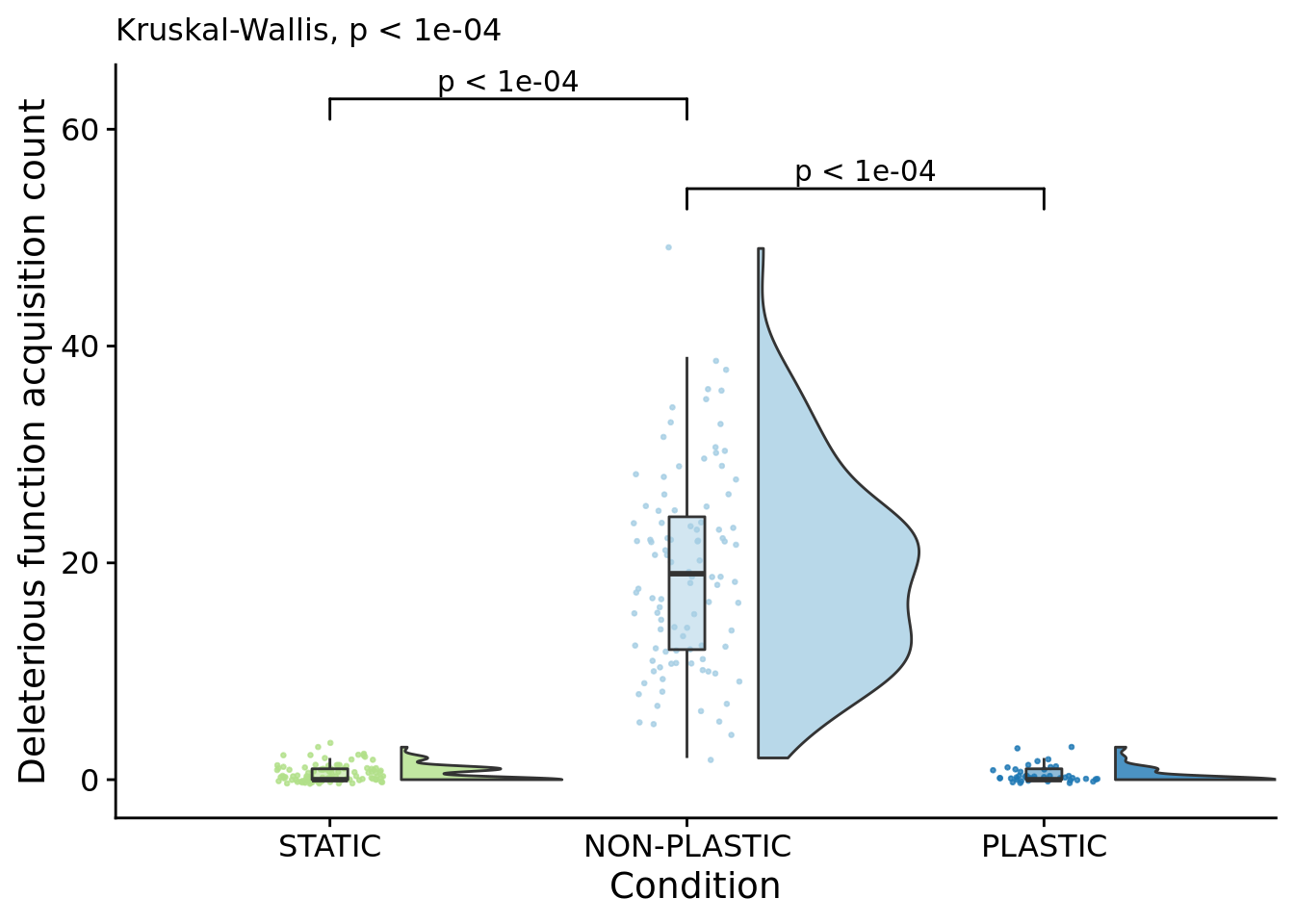
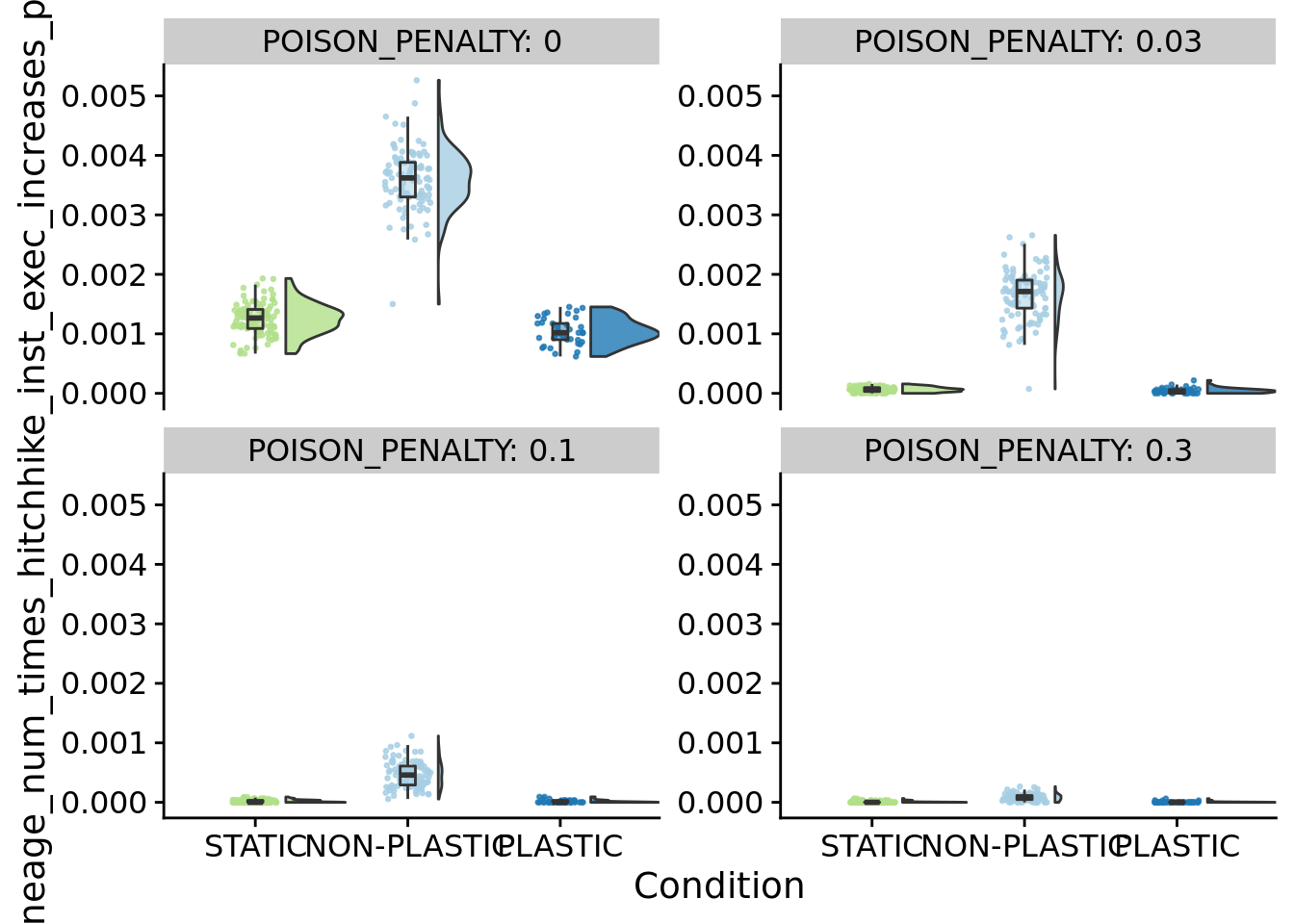
8.6.2 Frequency of increases in deleterious instruction execution (lineage)
ggplot(summary_data, aes(x=condition, y=dominant_lineage_num_times_hitchhike_inst_exec_increases_per_generation, fill=condition)) +
geom_flat_violin(
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Condition",
limits=condition_order
) +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
facet_wrap(
~POISON_PENALTY,
labeller=label_both,
scales="free_y"
) +
# coord_flip() +
theme(
legend.position="none"
)
ggsave(
paste0(working_directory, "plots/final-dominant-lineage-poison-increase-per-generation.png"),
width=15,
height=10
)for (penalty in poison_penalties) {
stat_data <- filter(summary_data, POISON_PENALTY==penalty)
print(
paste0(
"PENALTY: ", penalty
)
)
kt <- kruskal.test(
formula=dominant_lineage_num_times_hitchhike_inst_exec_increases_per_generation~condition,
data=stat_data
)
print(
kt
)
if (is.na(kt$p.value)) { next }
if (kt$p.value > 0.05) { next }
print(
pairwise.wilcox.test(
x=stat_data$dominant_lineage_num_times_hitchhike_inst_exec_increases_per_generation,
g=stat_data$condition,
p.adjust.method="bonferroni"
)
)
}## [1] "PENALTY: 0"
##
## Kruskal-Wallis rank sum test
##
## data: dominant_lineage_num_times_hitchhike_inst_exec_increases_per_generation by condition
## Kruskal-Wallis chi-squared = 180.05, df = 2, p-value < 2.2e-16
##
##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: stat_data$dominant_lineage_num_times_hitchhike_inst_exec_increases_per_generation and stat_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC < 2e-16 -
## STATIC < 2e-16 7.8e-05
##
## P value adjustment method: bonferroni
## [1] "PENALTY: 0.03"
##
## Kruskal-Wallis rank sum test
##
## data: dominant_lineage_num_times_hitchhike_inst_exec_increases_per_generation by condition
## Kruskal-Wallis chi-squared = 176.25, df = 2, p-value < 2.2e-16
##
##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: stat_data$dominant_lineage_num_times_hitchhike_inst_exec_increases_per_generation and stat_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC <2e-16 -
## STATIC <2e-16 0.019
##
## P value adjustment method: bonferroni
## [1] "PENALTY: 0.1"
##
## Kruskal-Wallis rank sum test
##
## data: dominant_lineage_num_times_hitchhike_inst_exec_increases_per_generation by condition
## Kruskal-Wallis chi-squared = 184.17, df = 2, p-value < 2.2e-16
##
##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: stat_data$dominant_lineage_num_times_hitchhike_inst_exec_increases_per_generation and stat_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC <2e-16 -
## STATIC <2e-16 0.2
##
## P value adjustment method: bonferroni
## [1] "PENALTY: 0.3"
##
## Kruskal-Wallis rank sum test
##
## data: dominant_lineage_num_times_hitchhike_inst_exec_increases_per_generation by condition
## Kruskal-Wallis chi-squared = 140.99, df = 2, p-value < 2.2e-16
##
##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: stat_data$dominant_lineage_num_times_hitchhike_inst_exec_increases_per_generation and stat_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC 2.2e-15 -
## STATIC < 2e-16 0.79
##
## P value adjustment method: bonferroniFigure for the manuscript:
# Compute manual labels for geom_signif
stat.test <- focal_summary_data %>%
wilcox_test(dominant_lineage_num_times_hitchhike_inst_exec_increases_per_generation ~ condition) %>%
adjust_pvalue(method = "bonferroni") %>%
add_significance() %>%
add_xy_position(x="condition", step.increase=0.2)
# Tweak y.position manually to account for scaled axis (edge case that triggers bad behavior in geom_signif)
stat.test$manual_position <- stat.test$y.position
stat.test$label <- mapply(p_label,stat.test$p.adj)
poison_increases_per_gen_fig <- ggplot(
focal_summary_data,
aes(x=condition, y=dominant_lineage_num_times_hitchhike_inst_exec_increases_per_generation, fill=condition)
) +
geom_flat_violin(
scale="width",
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Condition",
limits=condition_order,
labels=condition_order
) +
scale_y_continuous(
name="Deleterious function acquisition frequency",
) +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
# coord_flip()
labs(
subtitle=paste0(
"Kruskal-Wallis, ",
p_label(signif(kruskal.test(formula=dominant_lineage_num_times_hitchhike_inst_exec_increases_per_generation~condition, data=focal_summary_data)$p.value,digits=4))
)
) +
ggsignif::geom_signif(
data=filter(stat.test, p.adj <= alpha),
aes(xmin=group1,xmax=group2,annotations=label,y_position=manual_position),
manual=TRUE,
inherit.aes=FALSE
) +
theme(
legend.position="none"
)## Warning: Ignoring unknown aesthetics: xmin, xmax, annotations, y_position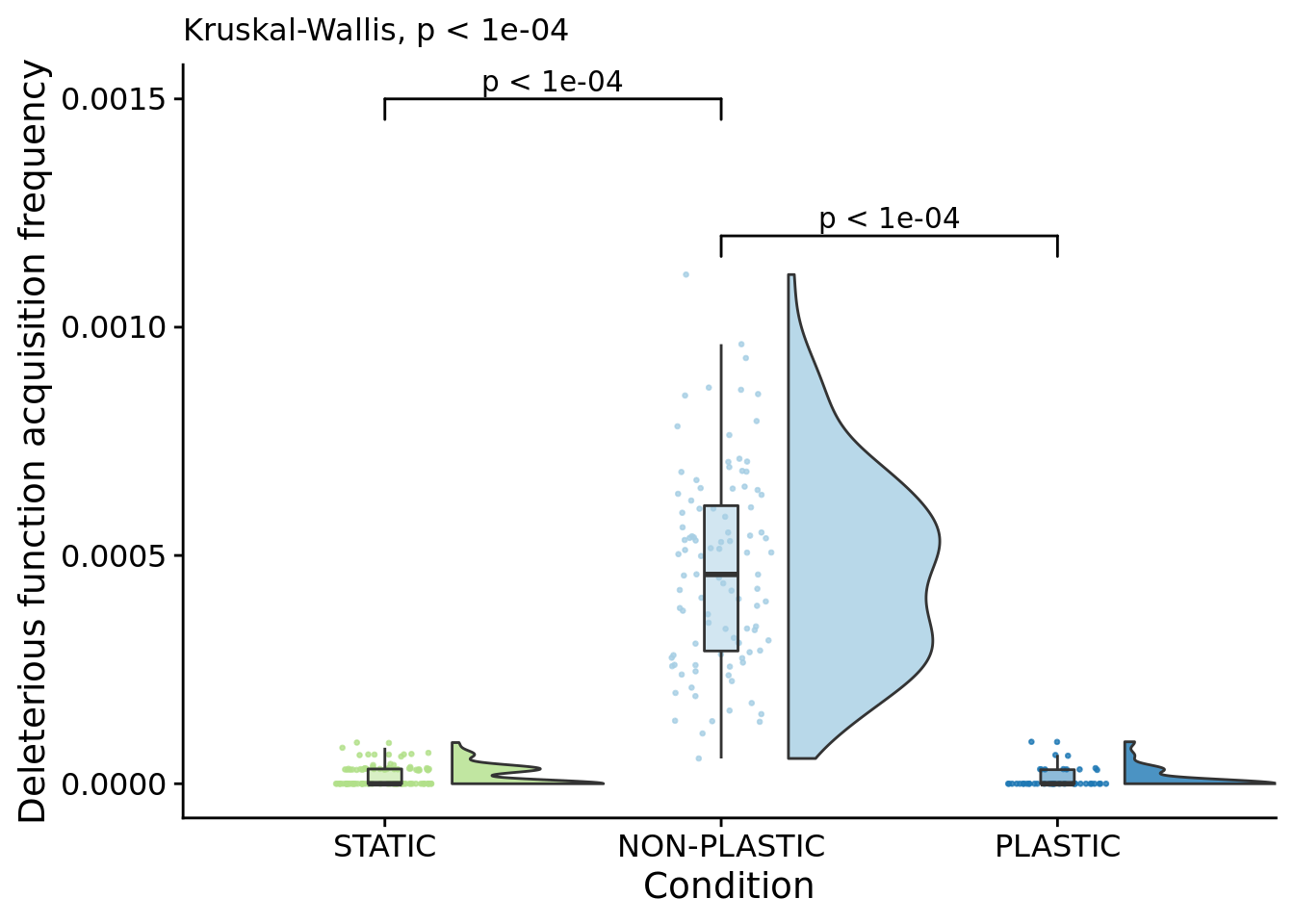
8.6.3 What fraction of mutations that increase deleterious instruction execution co-occur with base trait changes?
ggplot(filter(summary_data, dominant_lineage_num_times_hitchhike_inst_exec_increases>0), aes(x=frac_hitchhiking_linked_trait_change, fill=condition)) +
geom_density() +
facet_grid(
condition~POISON_PENALTY,
labeller=label_both,
scales="free_y"
) +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
theme(
legend.position="none"
)
ggsave(
paste0(working_directory, "plots/dominant-lineage-frac_hitchhiking_linked_trait_change.png"),
width=15,
height=10
)ggplot(filter(summary_data, dominant_lineage_num_times_hitchhike_inst_exec_increases>0 ), aes(x=condition, y=frac_hitchhiking_linked_trait_change, fill=condition)) +
geom_flat_violin(
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Condition",
limits=condition_order
) +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
facet_wrap(
~POISON_PENALTY,
labeller=label_both,
scales="free_y"
) +
# coord_flip() +
theme(
legend.position="none"
)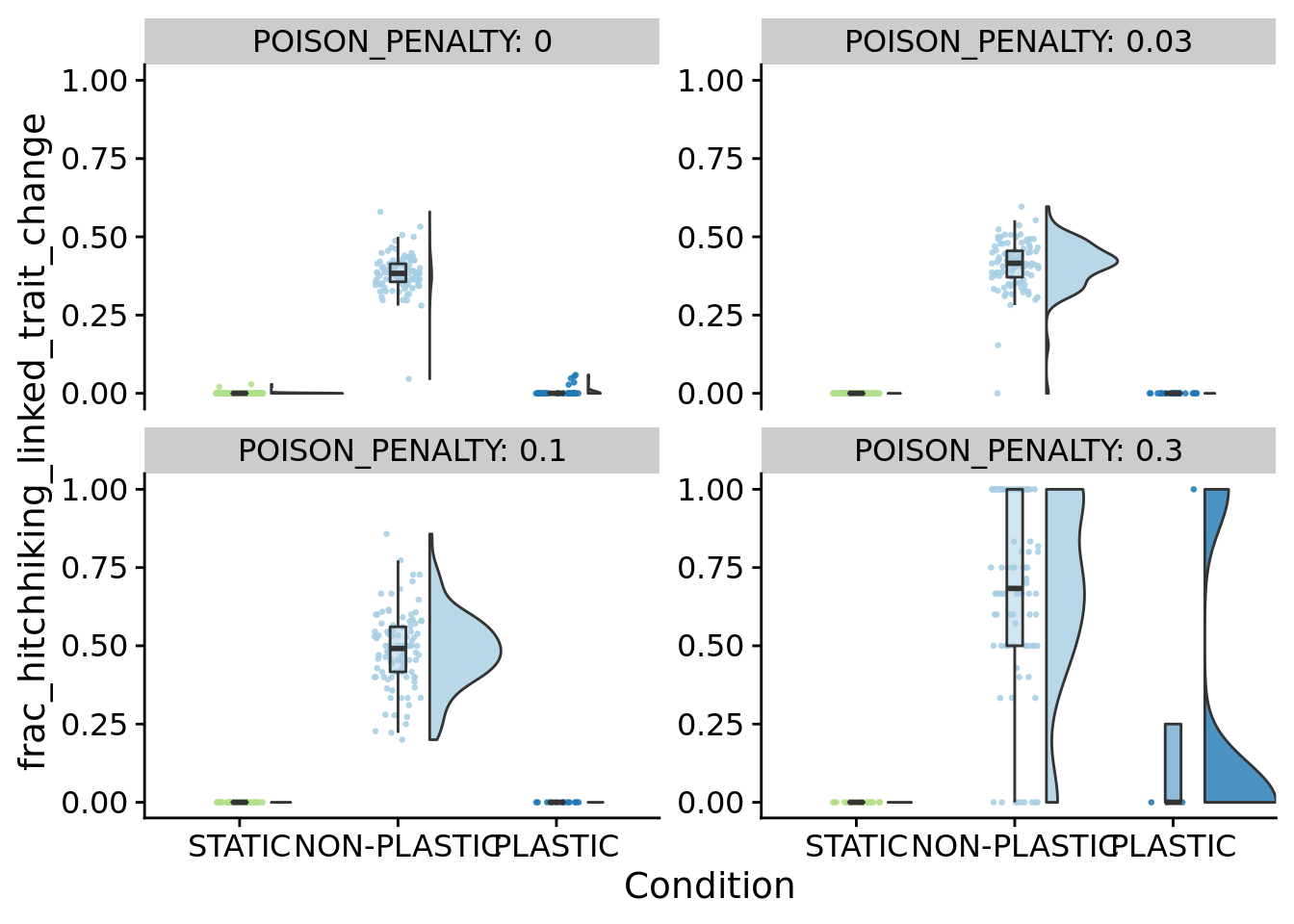
for (penalty in poison_penalties) {
stat_data <- filter(summary_data, POISON_PENALTY==penalty & dominant_lineage_num_times_hitchhike_inst_exec_increases>0)
print(
paste0(
"PENALTY: ", penalty
)
)
kt <- kruskal.test(
formula=frac_hitchhiking_linked_trait_change~condition,
data=stat_data
)
print(
kt
)
if (is.na(kt$p.value)) { next }
if (kt$p.value > 0.05) { next }
print(
pairwise.wilcox.test(
x=stat_data$frac_hitchhiking_linked_trait_change,
g=stat_data$condition,
p.adjust.method="bonferroni",
exact=FALSE
)
)
}## [1] "PENALTY: 0"
##
## Kruskal-Wallis rank sum test
##
## data: frac_hitchhiking_linked_trait_change by condition
## Kruskal-Wallis chi-squared = 211.29, df = 2, p-value < 2.2e-16
##
##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: stat_data$frac_hitchhiking_linked_trait_change and stat_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC <2e-16 -
## STATIC <2e-16 0.031
##
## P value adjustment method: bonferroni
## [1] "PENALTY: 0.03"
##
## Kruskal-Wallis rank sum test
##
## data: frac_hitchhiking_linked_trait_change by condition
## Kruskal-Wallis chi-squared = 186.88, df = 2, p-value < 2.2e-16
##
##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: stat_data$frac_hitchhiking_linked_trait_change and stat_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC 2.9e-16 -
## STATIC < 2e-16 -
##
## P value adjustment method: bonferroni
## [1] "PENALTY: 0.1"
##
## Kruskal-Wallis rank sum test
##
## data: frac_hitchhiking_linked_trait_change by condition
## Kruskal-Wallis chi-squared = 113.72, df = 2, p-value < 2.2e-16
##
##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: stat_data$frac_hitchhiking_linked_trait_change and stat_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC 3.3e-08 -
## STATIC < 2e-16 -
##
## P value adjustment method: bonferroni
## [1] "PENALTY: 0.3"
##
## Kruskal-Wallis rank sum test
##
## data: frac_hitchhiking_linked_trait_change by condition
## Kruskal-Wallis chi-squared = 34.791, df = 2, p-value = 2.788e-08
##
##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: stat_data$frac_hitchhiking_linked_trait_change and stat_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC 0.26 -
## STATIC 2.4e-08 0.18
##
## P value adjustment method: bonferronidenom <- sum(filter(summary_data, condition=="NON-PLASTIC" & POISON_PENALTY==0.1)$dominant_lineage_num_times_hitchhike_inst_exec_increases)
num <- sum(filter(summary_data, condition=="NON-PLASTIC" & POISON_PENALTY==0.1)$dominant_lineage_num_times_hitchhike_inst_exec_increases_with_primary_trait_change)
paste0("NON-PLASTIC (0.1 penalty): ", num/denom, "(", num, "/", denom, ")")## [1] "NON-PLASTIC (0.1 penalty): 0.498956158663883(956/1916)"denom <- sum(filter(summary_data, condition=="PLASTIC" & POISON_PENALTY==0.1)$dominant_lineage_num_times_hitchhike_inst_exec_increases)
num <- sum(filter(summary_data, condition=="PLASTIC" & POISON_PENALTY==0.1)$dominant_lineage_num_times_hitchhike_inst_exec_increases_with_primary_trait_change)
paste0("PLASTIC (0.1 penalty): ", num/denom, " (", num, "/", denom, ")")## [1] "PLASTIC (0.1 penalty): 0 (0/18)"denom <- sum(filter(summary_data, condition=="STATIC" & POISON_PENALTY==0.1)$dominant_lineage_num_times_hitchhike_inst_exec_increases)
num <- sum(filter(summary_data, condition=="STATIC" & POISON_PENALTY==0.1)$dominant_lineage_num_times_hitchhike_inst_exec_increases_with_primary_trait_change)
paste0("STATIC (0.1 penalty): ", num/denom, " (", num, "/", denom, ")")## [1] "STATIC (0.1 penalty): 0 (0/58)"Focal figure for the manuscript:
# Compute manual labels for geom_signif
stat.test <-filter(focal_summary_data,dominant_lineage_num_times_hitchhike_inst_exec_increases>0) %>%
wilcox_test(frac_hitchhiking_linked_trait_change ~ condition, comparisons=list(c("PLASTIC", "NON-PLASTIC"), c("STATIC", "NON-PLASTIC"))) %>%
adjust_pvalue(method = "bonferroni") %>%
add_significance() %>%
add_xy_position(x="condition")
# Tweak y.position manually to account for scaled axis (edge case that triggers bad behavior in geom_signif)
stat.test$manual_position <- stat.test$y.position
stat.test$label <- mapply(p_label,stat.test$p.adj)
linked_trait_change_fig <- ggplot(
filter(focal_summary_data, dominant_lineage_num_times_hitchhike_inst_exec_increases>0),
aes(x=condition, y=frac_hitchhiking_linked_trait_change, fill=condition)
) +
geom_flat_violin(
scale="width",
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Condition",
limits=condition_order,
labels=condition_order
) +
scale_y_continuous(
name="Fraction of linked deleterious function acquisition",
limits=c(-0.01, 1.2),
breaks=c(0, 0.25, 0.50, 0.75, 1.0)
) +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
labs(
subtitle=paste0(
"Kruskal-Wallis, ",
p_label(signif(kruskal.test(formula=frac_hitchhiking_linked_trait_change~condition, data=focal_summary_data)$p.value,digits=4))
)
) +
ggsignif::geom_signif(
data=filter(stat.test, p.adj <= alpha),
aes(xmin=group1,xmax=group2,annotations=label,y_position=manual_position),
manual=TRUE,
inherit.aes=FALSE
) +
theme(
legend.position="none"
)## Warning: Ignoring unknown aesthetics: xmin, xmax, annotations, y_position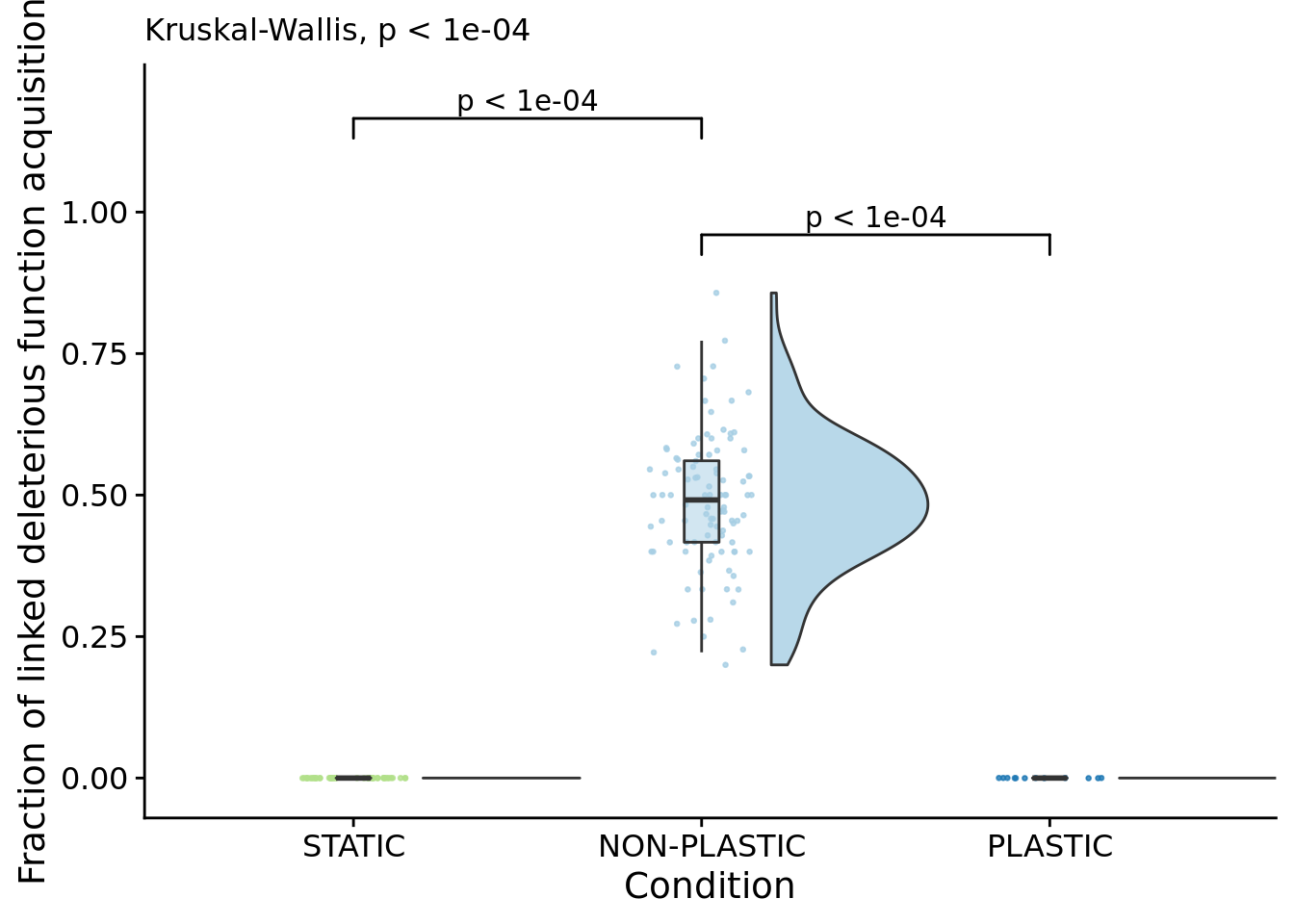
8.7 What fraction of deleterious execution increases occur in unexpressed phenotype (as cryptic variation)?
ggplot(filter(summary_data, dominant_lineage_num_times_hitchhike_inst_exec_increases>0 & condition=="PLASTIC"), aes(x=frac_unexpressed_hitchhiker_inc)) +
geom_density() +
facet_grid(
condition~POISON_PENALTY,
labeller=label_both,
scales="free_y"
) +
theme(
legend.position="none"
)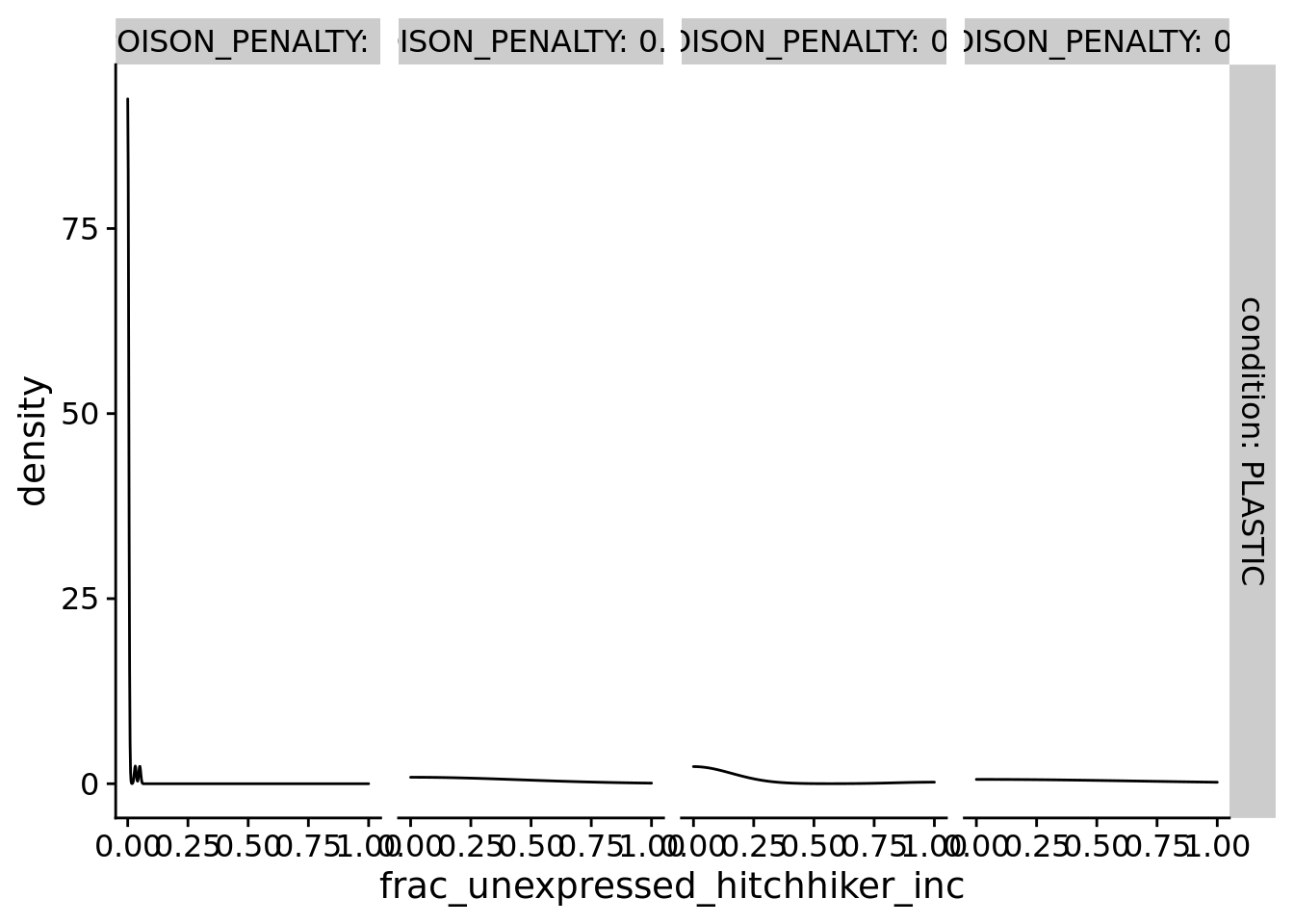
denom <- sum(filter(summary_data, condition=="PLASTIC" & POISON_PENALTY==0.1)$dominant_lineage_num_times_hitchhike_inst_exec_increases)
num <- sum(filter(summary_data, condition=="PLASTIC" & POISON_PENALTY==0.1)$dominant_lineage_num_times_hitchhike_inst_exec_increases_in_unexpressed_phenotype)
paste0("PLASTIC: ", num/denom, " (", num, "/", denom, ")")## [1] "PLASTIC: 0.0555555555555556 (1/18)"8.8 Manuscript figures
grid <- plot_grid(
poison_increases_fig +
theme(
axis.title.x=element_blank()
) +
ggtitle("Deleterious function acquisition count"),
poison_increases_per_gen_fig +
theme(
axis.title.x=element_blank()
) +
ggtitle("Deleterious function acquisition frequency"),
linked_trait_change_fig +
theme(
axis.title.x=element_blank()
) +
ggtitle("Linked deleterious function acquisition"),
nrow=1,
align="v",
labels="auto"
)
save_plot(
paste0(working_directory, "plots/", "poison-accumulation-panel.pdf"),
grid,
base_height=6,
base_asp=3/1
)
grid