Chapter 7 Evolution and maintenance of novel functions
The effect of adaptive phenotypic plasticity on the evolution and maintenance of novel functions.
7.1 Overview
total_updates <- 200000
replicates <- 100
alpha <- 0.05
focal_traits <- c("not","nand","and","ornot","or","andnot")
traits_set_a <- c("not", "and", "or")
traits_set_b <- c("nand", "ornot", "andnot")
extra_traits <- c(
"nor","xor","equals",
"logic_3aa","logic_3ab","logic_3ac",
"logic_3ad","logic_3ae","logic_3af",
"logic_3ag","logic_3ah","logic_3ai",
"logic_3aj","logic_3ak","logic_3al",
"logic_3am","logic_3an","logic_3ao",
"logic_3ap","logic_3aq","logic_3ar",
"logic_3as","logic_3at","logic_3au",
"logic_3av","logic_3aw","logic_3ax",
"logic_3ay","logic_3az","logic_3ba",
"logic_3bb","logic_3bc","logic_3bd",
"logic_3be","logic_3bf","logic_3bg",
"logic_3bh","logic_3bi","logic_3bj",
"logic_3bk","logic_3bl","logic_3bm",
"logic_3bn","logic_3bo","logic_3bp",
"logic_3bq","logic_3br","logic_3bs",
"logic_3bt","logic_3bu","logic_3bv",
"logic_3bw","logic_3bx","logic_3by",
"logic_3bz","logic_3ca","logic_3cb",
"logic_3cc","logic_3cd","logic_3ce",
"logic_3cf","logic_3cg","logic_3ch",
"logic_3ci","logic_3cj","logic_3ck",
"logic_3cl","logic_3cm","logic_3cn",
"logic_3co","logic_3cp"
)
# Relative location of data.
working_directory <- "experiments/2021-01-31-complex-features/analysis/" # << For bookdown
# working_directory <- "./"7.2 Analysis dependencies
Load all required R libraries.
library(ggplot2)
library(rstatix)
library(ggsignif)
library(scales)
library(tidyverse)
library(cowplot)
library(RColorBrewer)
library(Hmisc)
library(boot)
source("https://gist.githubusercontent.com/benmarwick/2a1bb0133ff568cbe28d/raw/fb53bd97121f7f9ce947837ef1a4c65a73bffb3f/geom_flat_violin.R")These analyses were conducted/knitted with the following computing environment:
## _
## platform x86_64-pc-linux-gnu
## arch x86_64
## os linux-gnu
## system x86_64, linux-gnu
## status
## major 4
## minor 1.3
## year 2022
## month 03
## day 10
## svn rev 81868
## language R
## version.string R version 4.1.3 (2022-03-10)
## nickname One Push-Up7.3 Setup
####### summary data #######
summary_data_loc <- paste0(working_directory, "data/aggregate.csv")
summary_data <- read.csv(summary_data_loc, na.strings="NONE")
summary_data$DISABLE_REACTION_SENSORS <- as.factor(summary_data$DISABLE_REACTION_SENSORS)
summary_data$chg_env <- summary_data$chg_env == "True"
summary_data$dominant_plastic_odd_even <- as.factor(summary_data$dominant_plastic_odd_even)
summary_data$sensors <- summary_data$DISABLE_REACTION_SENSORS == "0"
summary_data$is_plastic <- summary_data$dominant_plastic_odd_even == "True"
summary_data$extra_task_value <- as.factor(summary_data$extra_task_value)
summary_data <- filter(summary_data, extra_task_value == 0.1)
env_label_fun <- function(chg_env) {
if (chg_env) {
return("Fluctuating")
} else {
return("Constant")
}
}
sensors_label_fun <- function(has_sensors) {
if (has_sensors) {
return("Sensors")
} else {
return("No sensors")
}
}
condition_label_fun <- function(has_sensors, env_chg) {
if (has_sensors && env_chg) {
return("PLASTIC")
} else if (env_chg) {
return("NON-PLASTIC")
} else {
return("STATIC")
}
}
summary_data$env_label <- mapply(
env_label_fun,
summary_data$chg_env
)
summary_data$sensors_label <- mapply(
sensors_label_fun,
summary_data$sensors
)
summary_data$condition <- mapply(
condition_label_fun,
summary_data$sensors,
summary_data$chg_env
)
condition_order = c(
"STATIC",
"NON-PLASTIC",
"PLASTIC"
)
pairwise_comparisons <- list(
c("STATIC", "NON-PLASTIC"),
c("STATIC", "PLASTIC"),
c("PLASTIC", "NON-PLASTIC")
)
p_label <- function(p_value) {
threshold = 0.0001
if (p_value < threshold) {
return(paste0("p < ", threshold))
} else {
return(paste0("p = ", p_value))
}
}
# *really* inefficient way to identify outliers
is_outlier <- function(value, cond, data, column) {
cond_data <- filter(data, condition==cond)
q1 <- summary(cond_data[,column])[["1st Qu."]]
q3 <- summary(cond_data[,column])[["3rd Qu."]]
H <- 1.5 * IQR(cond_data[,column])
return( (value < (q1-H)) || (value > (q3+H)) )
}
###### time series #####
lineage_time_series_data_loc <- paste0(working_directory, "data/lineage_series.csv")
lineage_time_series_data <- read.csv(lineage_time_series_data_loc)
lineage_time_series_data$DISABLE_REACTION_SENSORS <- as.factor(lineage_time_series_data$DISABLE_REACTION_SENSORS)
lineage_time_series_data$chg_env <- lineage_time_series_data$chg_env == "True"
lineage_time_series_data$sensors <- lineage_time_series_data$DISABLE_REACTION_SENSORS == "0"
lineage_time_series_data$extra_task_value <- as.factor(lineage_time_series_data$extra_task_value)
lineage_time_series_data$env_label <- mapply(
env_label_fun,
lineage_time_series_data$chg_env
)
lineage_time_series_data$sensors_label <- mapply(
sensors_label_fun,
lineage_time_series_data$sensors
)
lineage_time_series_data$condition <- mapply(
condition_label_fun,
lineage_time_series_data$sensors,
lineage_time_series_data$chg_env
)
####### misc #######
# Configure our default graphing theme
theme_set(theme_cowplot())
# Palette
cb_palette <- "Paired"
# Create directory to dump plots
dir.create(paste0(working_directory, "plots"), showWarnings=FALSE)
# Sample mean function
samplemean <- function(x, d) {
return(mean(x[d]))
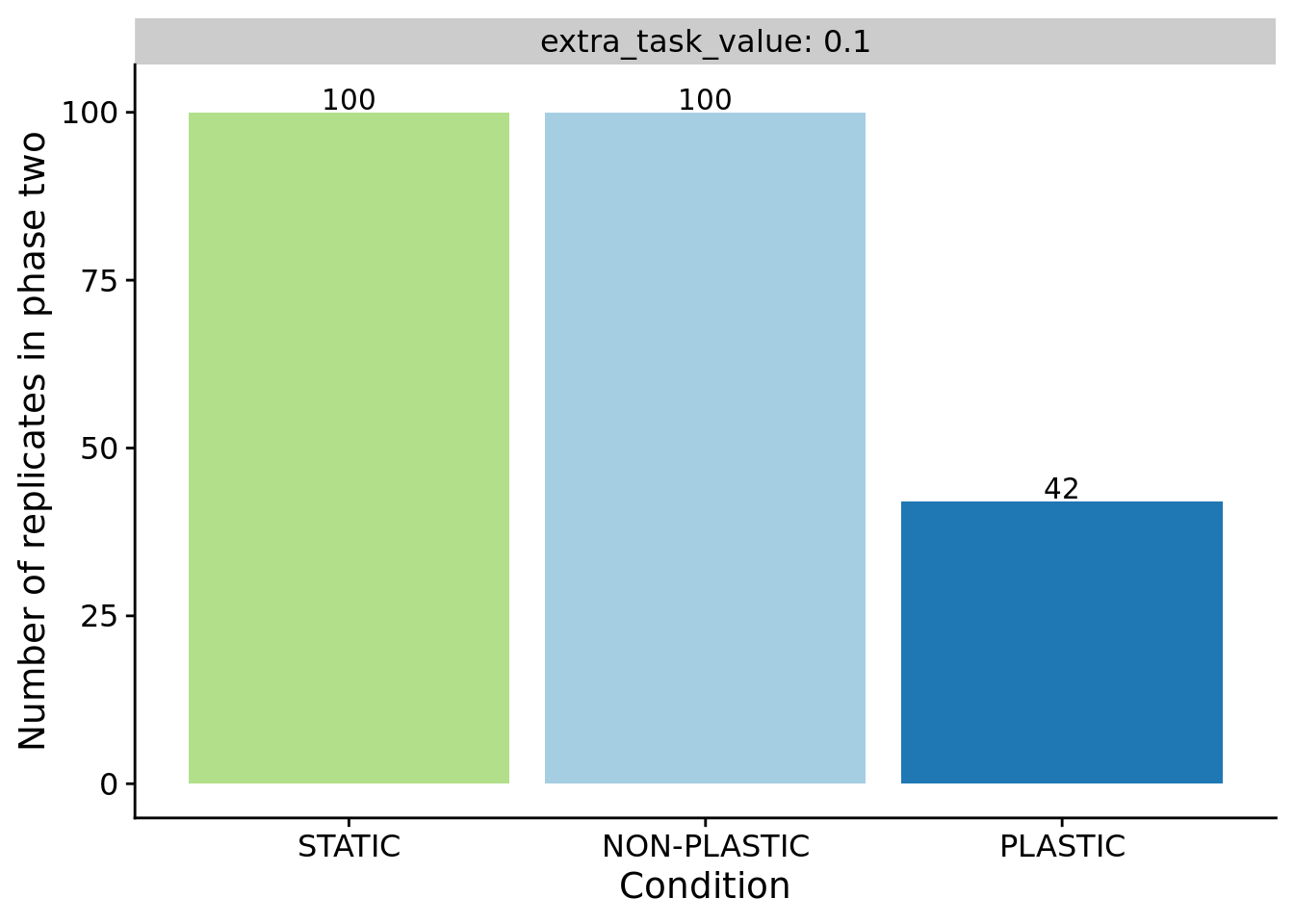
}7.4 The evolution of phenotypic plasticity
For sensor-enabled populations in fluctuating environments, we only transfered populations containing an optimally plastic genotype to phase two.
summary_data_grouped = dplyr::group_by(summary_data, sensors, env_label, condition, extra_task_value)
summary_data_group_counts = dplyr::summarize(summary_data_grouped, n=dplyr::n())
ggplot(summary_data_group_counts, aes(x=condition, y=n, fill=condition)) +
geom_col(position=position_dodge(0.9)) +
geom_text(aes(label=n, y=n+2)) +
scale_x_discrete(
name="Condition",
limits=condition_order
) +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
ylab("Number of replicates in phase two") +
facet_wrap(~extra_task_value, labeller=label_both) +
theme(
legend.position="none"
)
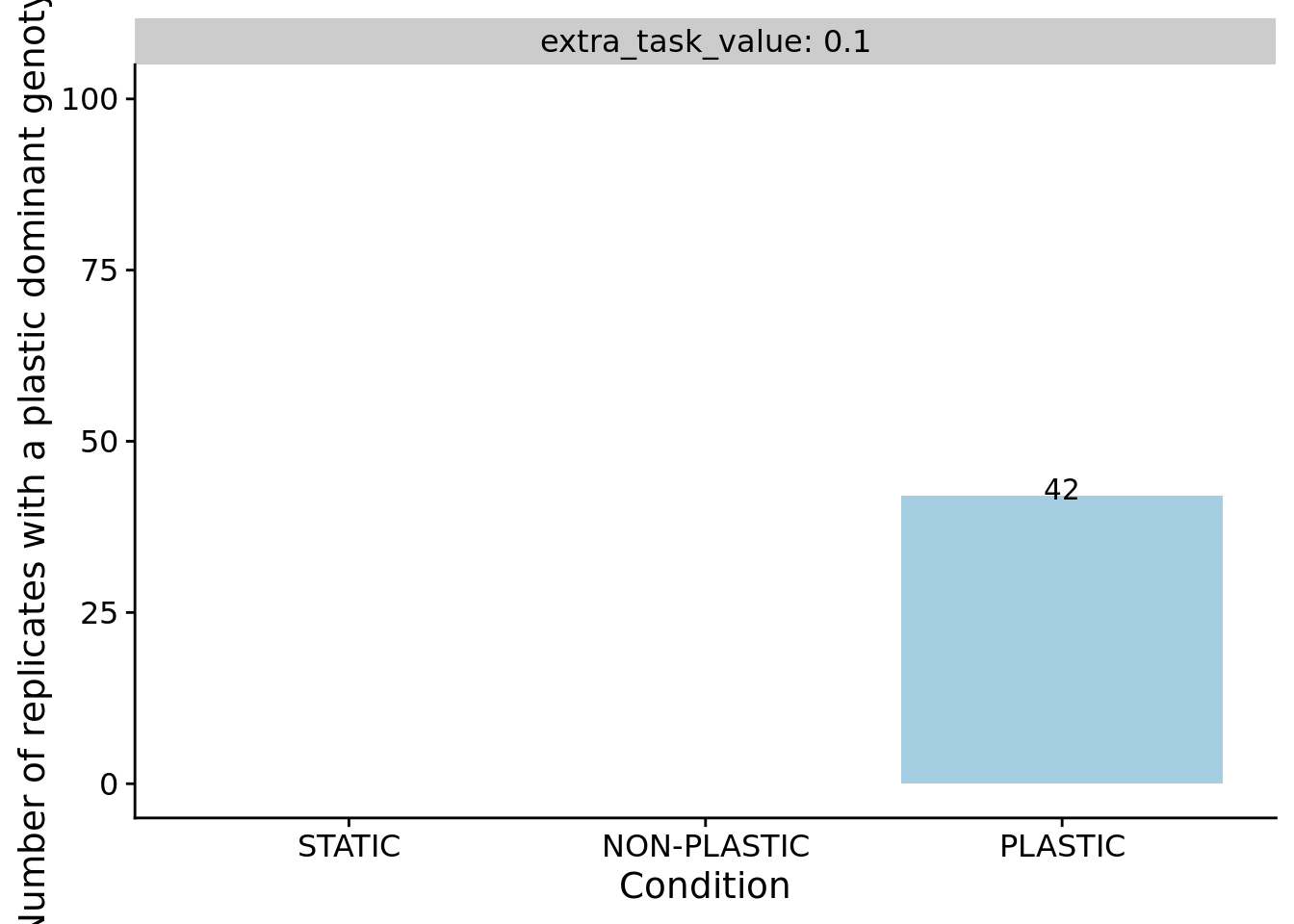
We can confirm our expectation that the dominant genotypes in non-plastic conditions are not phenotypically plastic.
summary_data_grouped = dplyr::group_by(summary_data, condition, is_plastic, extra_task_value)
summary_data_group_counts = dplyr::summarize(summary_data_grouped, n=dplyr::n())
ggplot(filter(summary_data_group_counts, is_plastic), aes(x=condition, y=n, fill=condition)) +
geom_col(position=position_dodge(0.9)) +
scale_x_discrete(
name="Condition",
limits=condition_order
) +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
ylim(0, 100) +
geom_text(aes(label=n, y=n+1)) +
ylab("Number of replicates with a plastic dominant genotype") +
facet_wrap(~extra_task_value, labeller=label_both) +
theme(
legend.position="none"
)
7.5 Final novel function count (dominant genotype)
How many novel functions do final dominant genotypes perform?
# Compute manual labels for geom_signif
stat.test <- summary_data %>%
wilcox_test(dominant_extra_tasks ~ condition) %>%
adjust_pvalue(method = "bonferroni") %>%
add_significance() %>%
add_xy_position(x="condition") # ,step.increase=1
# Tweak y.position manually to account for scaled axis (edge case that triggers bad behavior in geom_signif)
stat.test$manual_position <- stat.test$y.position #* c(1.0,1.0,1.03)
stat.test$label <- mapply(p_label,stat.test$p.adj)
summary_data$is_outlier <- mapply(
is_outlier,
summary_data$dominant_extra_tasks,
summary_data$condition,
MoreArgs=list(data=summary_data, column="dominant_extra_tasks")
)
final_novel_task_count_fig <- ggplot(
summary_data,
aes(x=condition, y=dominant_extra_tasks, fill=condition)
) +
geom_flat_violin(
# data=filter(summary_data,is_outlier==FALSE),
scale="width",
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Condition",
limits=condition_order,
labels=condition_order
) +
scale_y_continuous(
name="Final novel function count"
) +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
labs(
subtitle=paste0(
"Kruskal-Wallis, ",
p_label(signif(kruskal.test(formula=dominant_extra_tasks~condition, data=summary_data)$p.value,digits=4))
)
) +
ggsignif::geom_signif(
data=filter(stat.test, p.adj <= alpha),
aes(xmin=group1,xmax=group2,annotations=label,y_position=manual_position),
manual=TRUE,
inherit.aes=FALSE
) +
# coord_flip()
theme(
legend.position="none"
)## Warning: Ignoring unknown aesthetics: xmin, xmax, annotations, y_position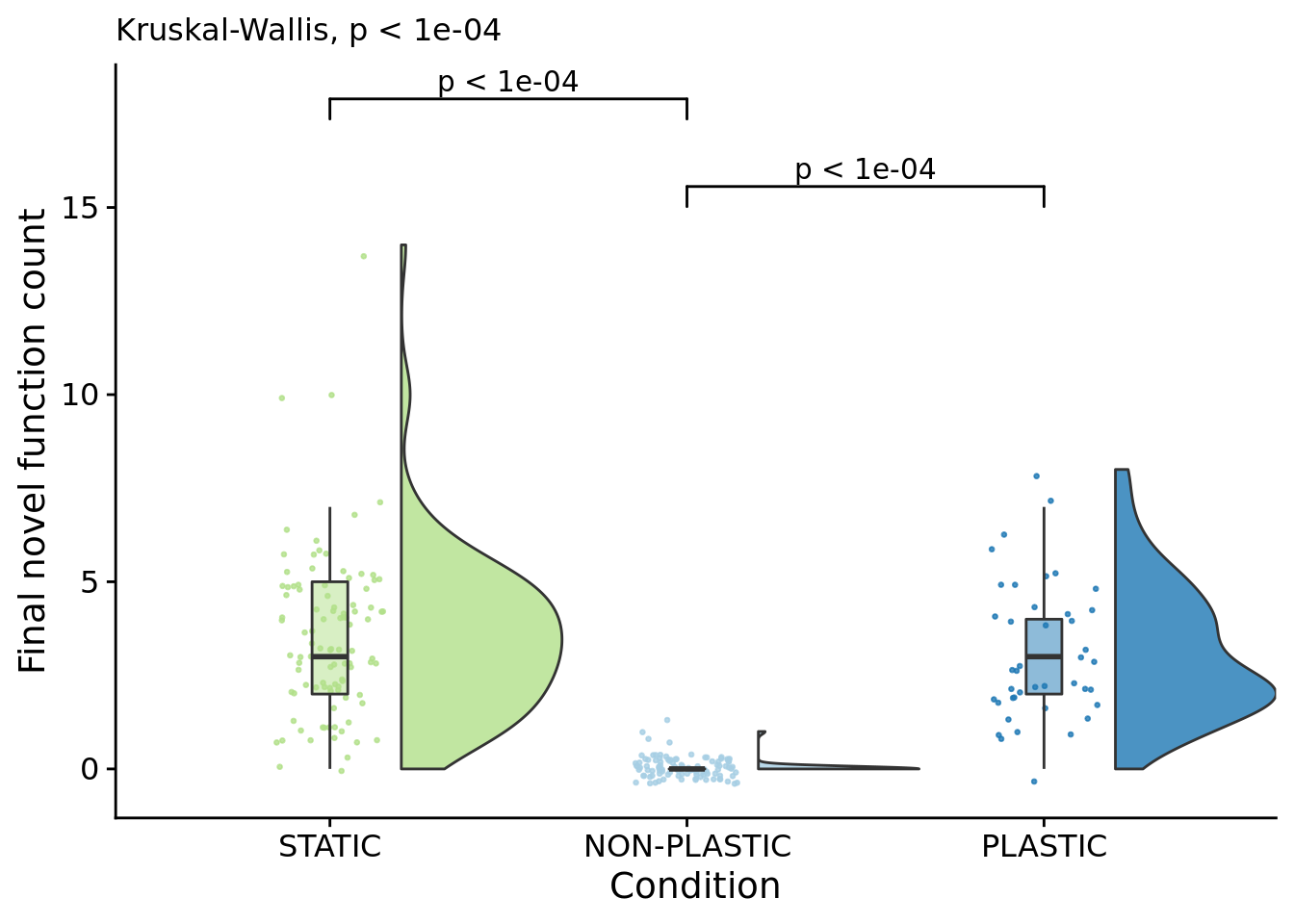
##
## Kruskal-Wallis rank sum test
##
## data: dominant_extra_tasks by condition
## Kruskal-Wallis chi-squared = 177.17, df = 2, p-value < 2.2e-16pairwise.wilcox.test(
x=summary_data$dominant_extra_tasks,
g=summary_data$condition,
p.adjust.method="bonferroni",
conf.int=TRUE,
conf.level=0.95
)##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: summary_data$dominant_extra_tasks and summary_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC <2e-16 -
## STATIC <2e-16 0.9
##
## P value adjustment method: bonferronipaste(
sep="; ",
paste0(
"PLASTIC median: ",
median(filter(summary_data, condition=="PLASTIC")$dominant_extra_tasks)
),
paste0(
"STATIC median: ",
median(filter(summary_data, condition=="STATIC")$dominant_extra_tasks)
),
paste0(
"NON-PLASTIC median: ",
median(filter(summary_data, condition=="NON-PLASTIC")$dominant_extra_tasks)
)
)## [1] "PLASTIC median: 3; STATIC median: 3; NON-PLASTIC median: 0"## [1] "Wilcox rank sum test statistics:"for (pair in pairwise_comparisons) {
pair_data <- filter(summary_data, condition %in% pair)
pair_data$condition <- as.factor(pair_data$condition)
wt <- wilcox.test(
formula=dominant_extra_tasks~condition,
data=pair_data,
exact=FALSE,
paired=FALSE
)
print(paste0(pair[1], "<-->", pair[2], ": W=",wt$statistic))
}## [1] "STATIC<-->NON-PLASTIC: W=184"
## [1] "STATIC<-->PLASTIC: W=1871"
## [1] "PLASTIC<-->NON-PLASTIC: W=64"7.6 Novel function count (final population)
How many novel functions are performed across the final population (1% of organisms must perform to count)?
ggplot(summary_data, aes(x=condition, y=final_pop_extra_tasks_0.01, fill=condition)) +
geom_flat_violin(
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Condition",
limits=condition_order
) +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
# coord_flip() +
theme(
legend.position="none"
)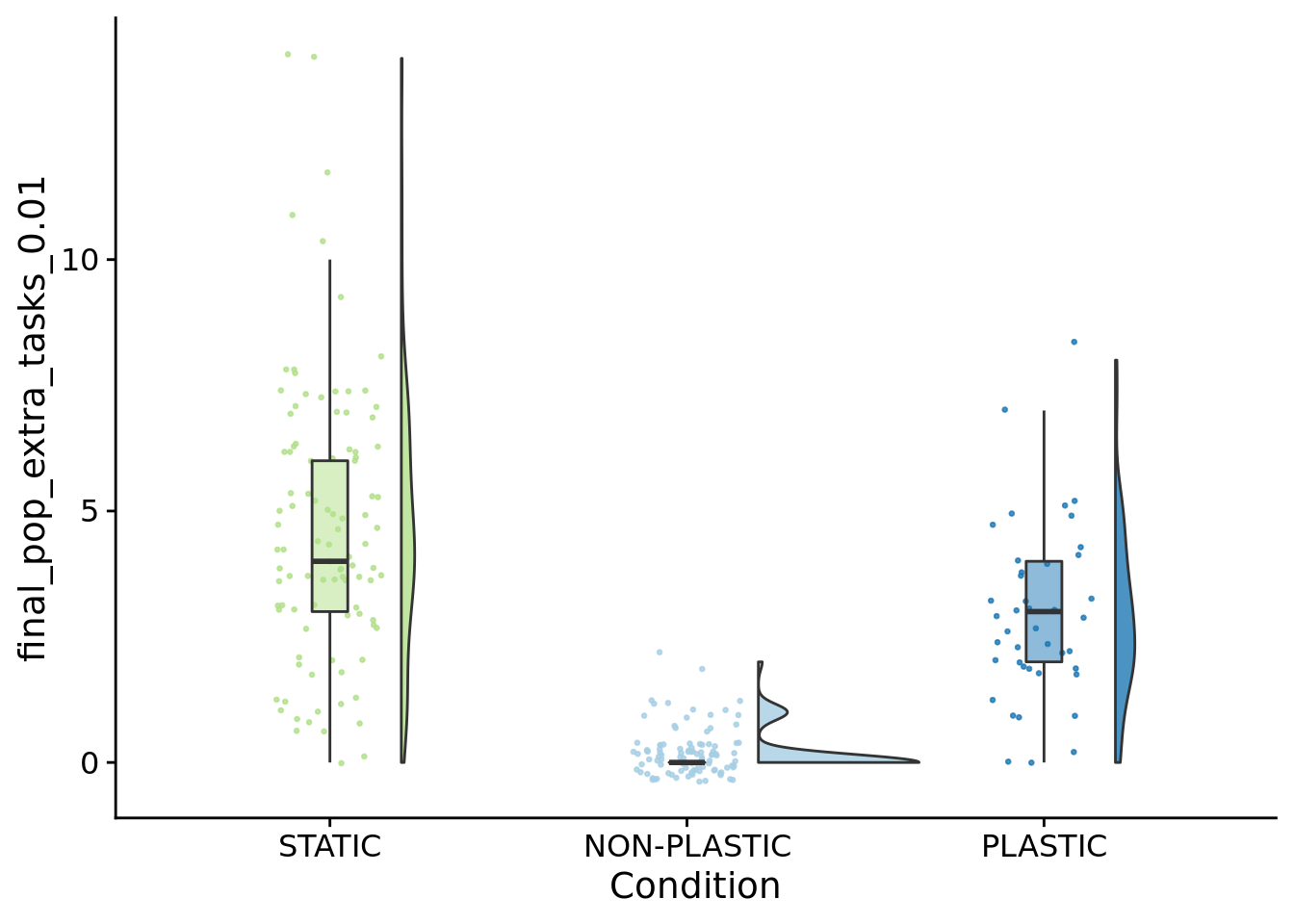
##
## Kruskal-Wallis rank sum test
##
## data: final_pop_extra_tasks_0.01 by condition
## Kruskal-Wallis chi-squared = 169.47, df = 2, p-value < 2.2e-16pairwise.wilcox.test(
x=summary_data$final_pop_extra_tasks_0.01,
g=summary_data$condition,
p.adjust.method="bonferroni",
conf.int=TRUE,
conf.level=0.95
)##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: summary_data$final_pop_extra_tasks_0.01 and summary_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC < 2e-16 -
## STATIC < 2e-16 0.00016
##
## P value adjustment method: bonferronipaste(
sep="; ",
paste0(
"PLASTIC median: ",
median(filter(summary_data, condition=="PLASTIC")$final_pop_extra_tasks_0.01)
),
paste0(
"STATIC median: ",
median(filter(summary_data, condition=="STATIC")$final_pop_extra_tasks_0.01)
),
paste0(
"NON-PLASTIC median: ",
median(filter(summary_data, condition=="NON-PLASTIC")$final_pop_extra_tasks_0.01)
)
)## [1] "PLASTIC median: 3; STATIC median: 4; NON-PLASTIC median: 0"## [1] "Wilcox rank sum test statistics:"for (pair in pairwise_comparisons) {
pair_data <- filter(summary_data, condition %in% pair)
pair_data$condition <- as.factor(pair_data$condition)
wt <- wilcox.test(
formula=final_pop_extra_tasks_0.01~condition,
data=pair_data,
exact=FALSE,
paired=FALSE
)
print(paste0(pair[1], "<-->", pair[2], ": W=",wt$statistic))
}## [1] "STATIC<-->NON-PLASTIC: W=227.5"
## [1] "STATIC<-->PLASTIC: W=1203"
## [1] "PLASTIC<-->NON-PLASTIC: W=225.5"7.7 Novel function discovery (lineage)
# Compute manual labels for geom_signif
stat.test <- summary_data %>%
wilcox_test(dominant_lineage_extra_traits_discovered ~ condition) %>%
adjust_pvalue(method = "bonferroni") %>%
add_significance() %>%
add_xy_position(x="condition") # ,step.increase=1
# Tweak y.position manually to account for scaled axis (edge case that triggers bad behavior in geom_signif)
stat.test$manual_position <- stat.test$y.position #* c(1.0,1.0,1.03)
stat.test$label <- mapply(p_label,stat.test$p.adj)
summary_data$is_outlier <- mapply(
is_outlier,
summary_data$dominant_lineage_extra_traits_discovered,
summary_data$condition,
MoreArgs=list(data=summary_data, column="dominant_lineage_extra_traits_discovered")
)
lineage_novel_task_discovery_fig <- ggplot(
summary_data,
aes(x=condition, y=dominant_lineage_extra_traits_discovered, fill=condition)
) +
geom_flat_violin(
# data=filter(summary_data,is_outlier==FALSE),
scale="width",
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Condition",
limits=condition_order,
labels=condition_order
) +
scale_y_continuous(
name="Novel function discovery"
) +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
labs(
subtitle=paste0(
"Kruskal-Wallis, ",
p_label(signif(kruskal.test(formula=dominant_lineage_extra_traits_discovered~condition, data=summary_data)$p.value,digits=4))
)
) +
ggsignif::geom_signif(
data=filter(stat.test, p.adj <= alpha),
aes(xmin=group1,xmax=group2,annotations=label,y_position=manual_position),
manual=TRUE,
inherit.aes=FALSE
) +
# coord_flip()
theme(
legend.position="none"
)## Warning: Ignoring unknown aesthetics: xmin, xmax, annotations, y_position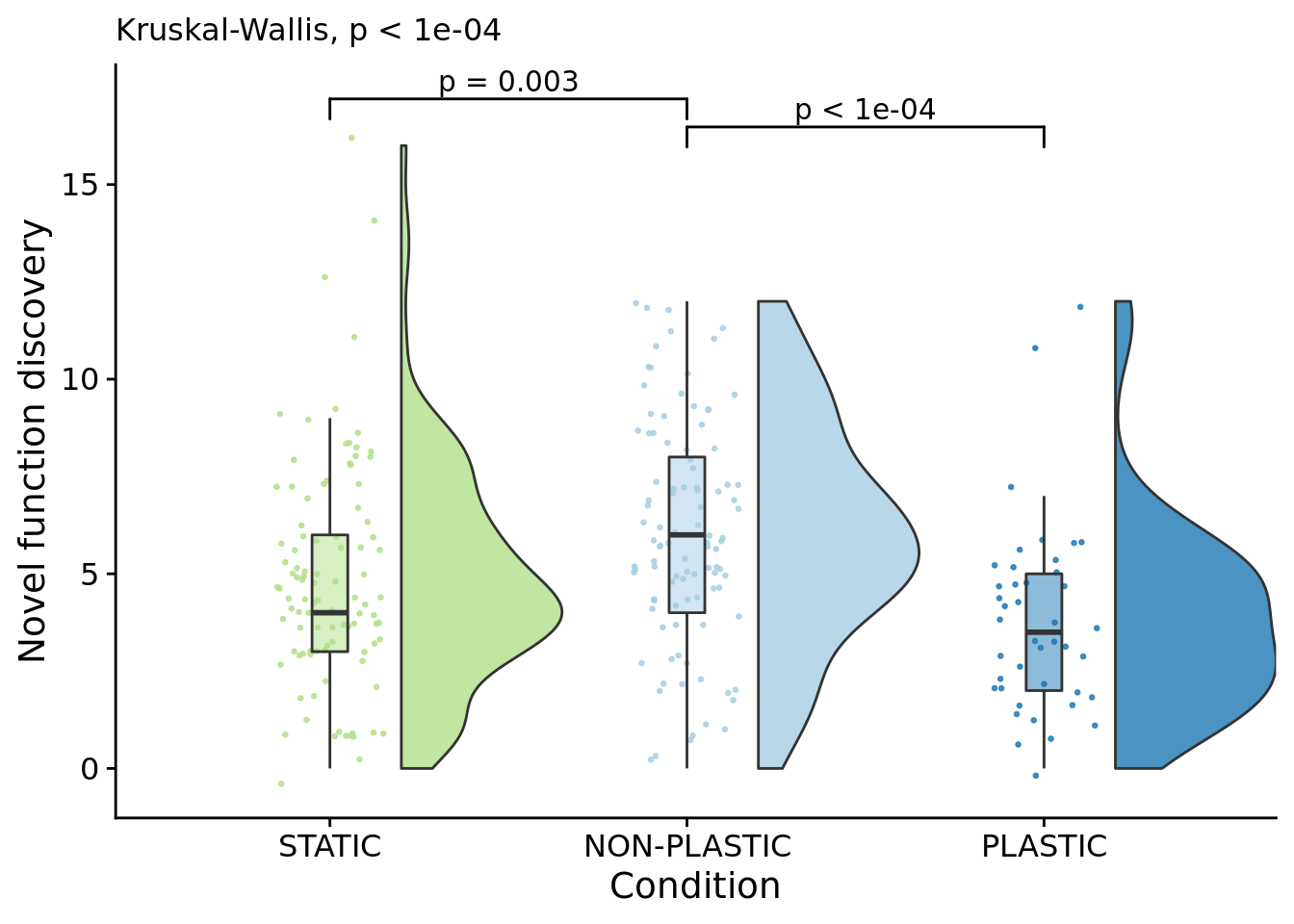
##
## Kruskal-Wallis rank sum test
##
## data: dominant_lineage_extra_traits_discovered by condition
## Kruskal-Wallis chi-squared = 24.099, df = 2, p-value = 5.846e-06pairwise.wilcox.test(
x=summary_data$dominant_lineage_extra_traits_discovered,
g=summary_data$condition,
p.adjust.method="bonferroni",
conf.int=TRUE,
conf.level=0.95
)##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: summary_data$dominant_lineage_extra_traits_discovered and summary_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC 1.7e-05 -
## STATIC 0.0035 0.0561
##
## P value adjustment method: bonferronipaste(
sep="; ",
paste0(
"PLASTIC median: ",
median(filter(summary_data, condition=="PLASTIC")$dominant_lineage_extra_traits_discovered)
),
paste0(
"STATIC median: ",
median(filter(summary_data, condition=="STATIC")$dominant_lineage_extra_traits_discovered)
),
paste0(
"NON-PLASTIC median: ",
median(filter(summary_data, condition=="NON-PLASTIC")$dominant_lineage_extra_traits_discovered)
)
)## [1] "PLASTIC median: 3.5; STATIC median: 4; NON-PLASTIC median: 6"## [1] "Wilcox rank sum test statistics:"for (pair in pairwise_comparisons) {
pair_data <- filter(summary_data, condition %in% pair)
pair_data$condition <- as.factor(pair_data$condition)
wt <- wilcox.test(
formula=dominant_lineage_extra_traits_discovered~condition,
data=pair_data,
exact=FALSE,
paired=FALSE
)
print(paste0(pair[1], "<-->", pair[2], ": W=",wt$statistic))
}## [1] "STATIC<-->NON-PLASTIC: W=6319.5"
## [1] "STATIC<-->PLASTIC: W=1578"
## [1] "PLASTIC<-->NON-PLASTIC: W=3110.5"7.8 Novel function discovery (population)
ggplot(
summary_data,
aes(x=condition, y=discovered_extra_tasks_0.01, fill=condition)
) +
geom_flat_violin(
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Condition",
limits=condition_order
) +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
# coord_flip() +
theme(
legend.position="none"
)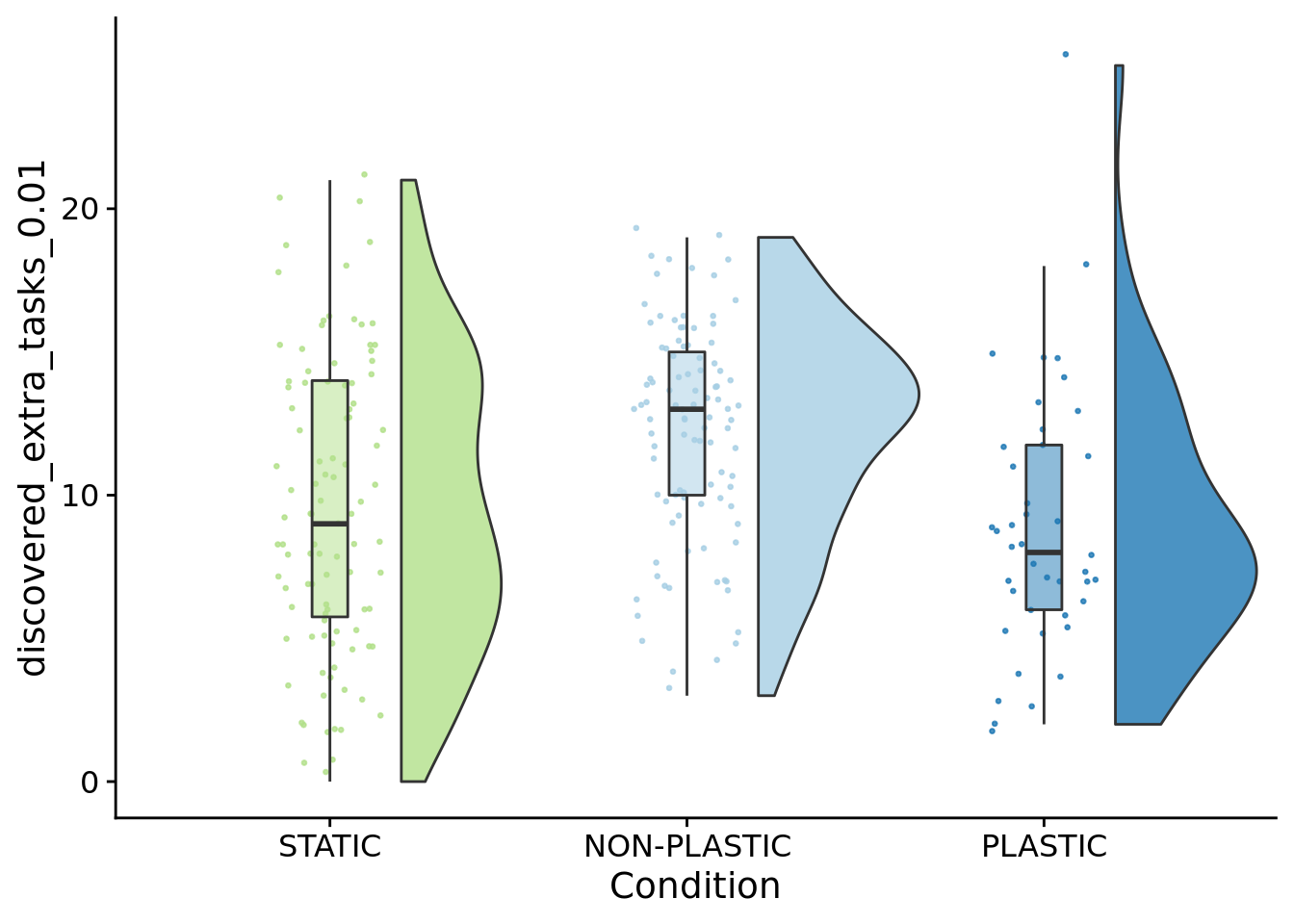
##
## Kruskal-Wallis rank sum test
##
## data: discovered_extra_tasks_0.01 by condition
## Kruskal-Wallis chi-squared = 24.271, df = 2, p-value = 5.365e-06pairwise.wilcox.test(
x=summary_data$discovered_extra_tasks_0.01,
g=summary_data$condition,
p.adjust.method="bonferroni",
conf.int=TRUE,
conf.level=0.95
)##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: summary_data$discovered_extra_tasks_0.01 and summary_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC 2.4e-05 -
## STATIC 0.00035 1.00000
##
## P value adjustment method: bonferronipaste(
sep="; ",
paste0(
"PLASTIC median: ",
median(filter(summary_data, condition=="PLASTIC")$discovered_extra_tasks_0.01)
),
paste0(
"STATIC median: ",
median(filter(summary_data, condition=="STATIC")$discovered_extra_tasks_0.01)
),
paste0(
"NON-PLASTIC median: ",
median(filter(summary_data, condition=="NON-PLASTIC")$discovered_extra_tasks_0.01)
)
)## [1] "PLASTIC median: 8; STATIC median: 9; NON-PLASTIC median: 13"## [1] "Wilcox rank sum test statistics:"for (pair in pairwise_comparisons) {
pair_data <- filter(summary_data, condition %in% pair)
pair_data$condition <- as.factor(pair_data$condition)
wt <- wilcox.test(
formula=discovered_extra_tasks_0.01~condition,
data=pair_data,
exact=FALSE,
paired=FALSE
)
print(paste0(pair[1], "<-->", pair[2], ": W=",wt$statistic))
}## [1] "STATIC<-->NON-PLASTIC: W=6573.5"
## [1] "STATIC<-->PLASTIC: W=1918.5"
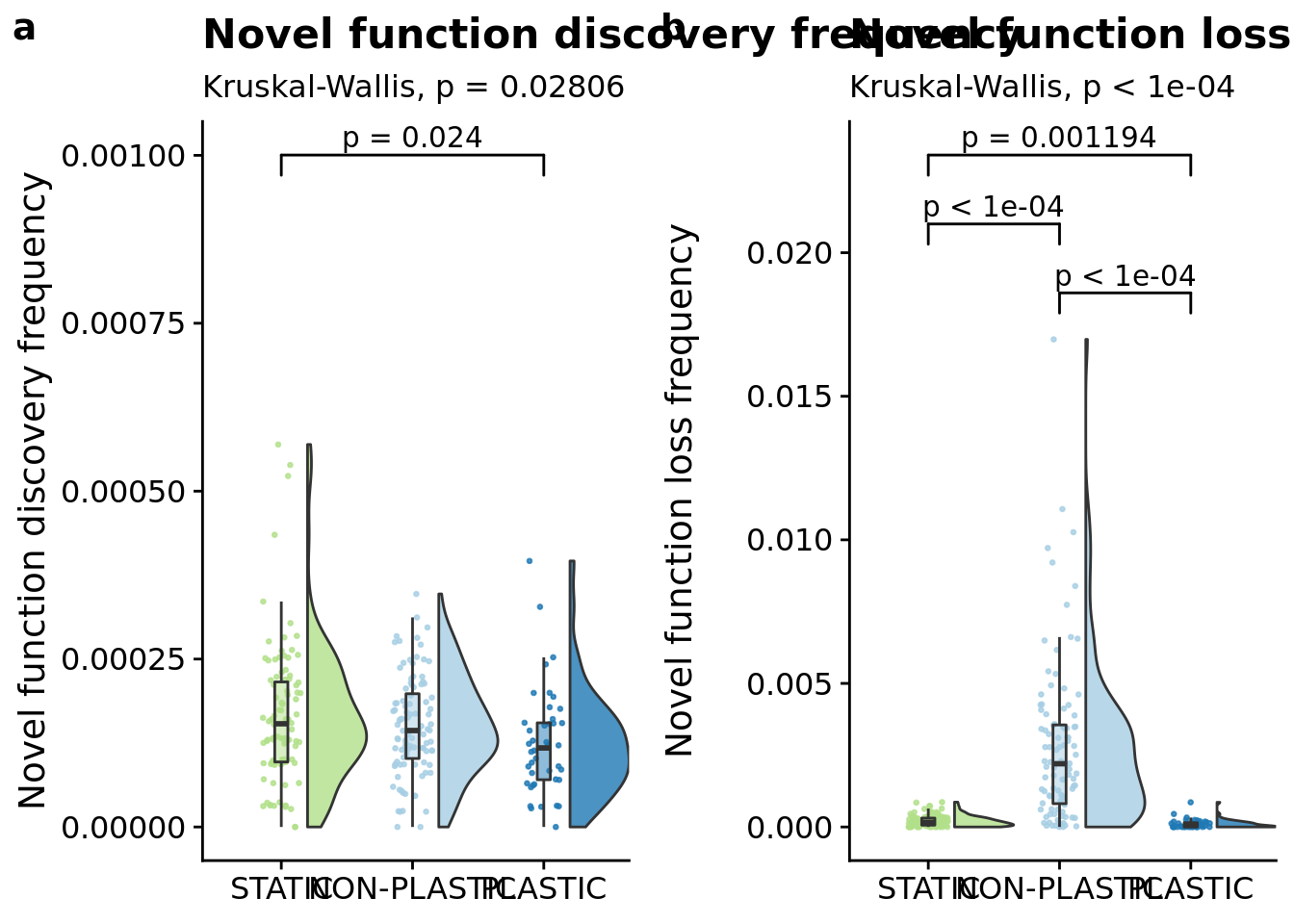
## [1] "PLASTIC<-->NON-PLASTIC: W=3096"7.9 Novel function discovery frequency (lineage)
summary_data$dominant_lineage_extra_traits_discovered_per_generation <- summary_data$dominant_lineage_extra_traits_discovered / summary_data$dominant_generation_born
summary_data$dominant_lineage_extra_traits_generations_per_discovery <- summary_data$dominant_generation_born / summary_data$dominant_lineage_extra_traits_discovered
# Compute manual labels for geom_signif
# stat.test <- filter(summary_data, dominant_lineage_extra_traits_discovered > 0) %>%
# wilcox_test(dominant_lineage_extra_traits_generations_per_discovery ~ condition) %>%
# adjust_pvalue(method = "bonferroni") %>%
# add_significance() %>%
# add_xy_position(x="condition") # ,step.increase=1
stat.test <- summary_data %>%
wilcox_test(dominant_lineage_extra_traits_discovered_per_generation ~ condition) %>%
adjust_pvalue(method = "bonferroni") %>%
add_significance() %>%
add_xy_position(x="condition", step.increase=0.0001) # ,step.increase=1
# Tweak y.position manually to account for scaled axis (edge case that triggers bad behavior in geom_signif)
stat.test$manual_position <- stat.test$y.position #* c(1.0,1.0,1.03)
stat.test$label <- mapply(p_label,stat.test$p.adj)
summary_data$is_outlier <- mapply(
is_outlier,
summary_data$dominant_lineage_extra_traits_discovered_per_generation,
summary_data$condition,
MoreArgs=list(data=summary_data, column="dominant_lineage_extra_traits_discovered_per_generation")
)
lineage_novel_task_discovery_freq_fig <- ggplot(
# filter(summary_data, dominant_lineage_extra_traits_discovered > 0),
summary_data,
aes(x=condition, y=dominant_lineage_extra_traits_discovered_per_generation, fill=condition)
) +
geom_flat_violin(
# data=filter(summary_data,is_outlier==FALSE),
scale="width",
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Condition",
limits=condition_order
) +
ylab("Novel function discovery frequency") +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
labs(
subtitle=paste0(
"Kruskal-Wallis, ",
p_label(signif(kruskal.test(formula=dominant_lineage_extra_traits_discovered_per_generation~condition, data=summary_data)$p.value,digits=4))
)
) +
ggsignif::geom_signif(
data=filter(stat.test, p.adj <= alpha),
aes(xmin=group1,xmax=group2,annotations=label,y_position=manual_position),
manual=TRUE,
inherit.aes=FALSE
) +
# coord_flip() +
theme(
legend.position="none"
)## Warning: Ignoring unknown aesthetics: xmin, xmax, annotations, y_position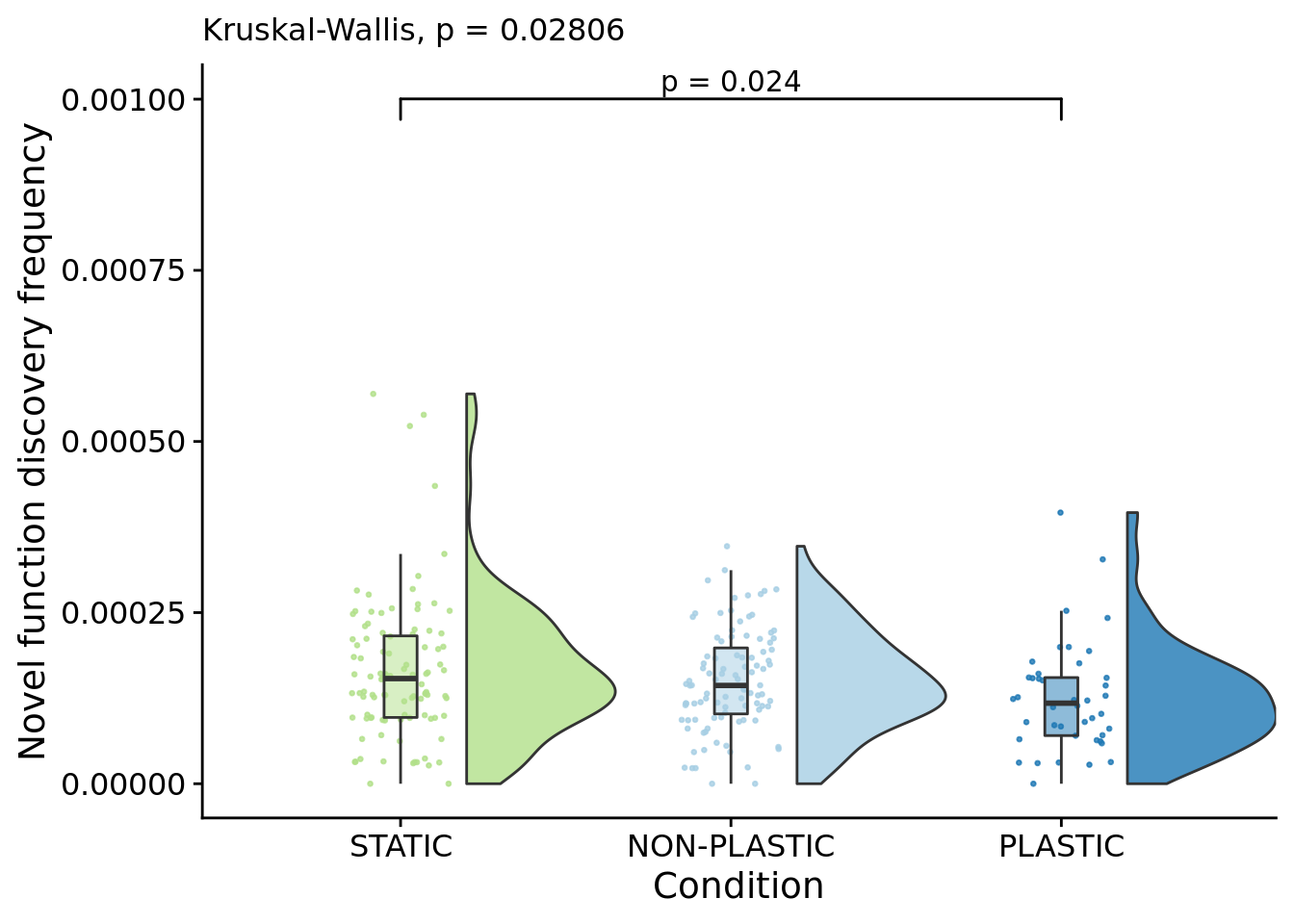
kruskal.test(
formula=dominant_lineage_extra_traits_discovered_per_generation~condition,
data=summary_data
)##
## Kruskal-Wallis rank sum test
##
## data: dominant_lineage_extra_traits_discovered_per_generation by condition
## Kruskal-Wallis chi-squared = 7.1465, df = 2, p-value = 0.02806pairwise.wilcox.test(
x=summary_data$dominant_lineage_extra_traits_discovered_per_generation,
g=summary_data$condition,
p.adjust.method="bonferroni",
conf.int=TRUE,
conf.level=0.95
)##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: summary_data$dominant_lineage_extra_traits_discovered_per_generation and summary_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC 0.092 -
## STATIC 1.000 0.025
##
## P value adjustment method: bonferronipaste(
sep="; ",
paste0(
"PLASTIC median: ",
median(filter(summary_data, condition=="PLASTIC")$dominant_lineage_extra_traits_discovered_per_generation)
),
paste0(
"STATIC median: ",
median(filter(summary_data, condition=="STATIC")$dominant_lineage_extra_traits_discovered_per_generation)
),
paste0(
"NON-PLASTIC median: ",
median(filter(summary_data, condition=="NON-PLASTIC")$dominant_lineage_extra_traits_discovered_per_generation)
)
)## [1] "PLASTIC median: 0.000117695011124939; STATIC median: 0.00015363220504867; NON-PLASTIC median: 0.00014358046266055"## [1] "Wilcox rank sum test statistics:"for (pair in pairwise_comparisons) {
pair_data <- filter(summary_data, condition %in% pair)
pair_data$condition <- as.factor(pair_data$condition)
wt <- wilcox.test(
formula=dominant_lineage_extra_traits_discovered_per_generation~condition,
data=pair_data,
exact=FALSE,
paired=FALSE
)
print(paste0(pair[1], "<-->", pair[2], ": W=",wt$statistic))
}## [1] "STATIC<-->NON-PLASTIC: W=4751"
## [1] "STATIC<-->PLASTIC: W=1510.5"
## [1] "PLASTIC<-->NON-PLASTIC: W=2584"7.10 Novel functions gained (lineage)
ggplot(
summary_data,
aes(x=condition, y=dominant_lineage_extra_traits_gained, fill=condition)
) +
geom_flat_violin(
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Condition",
limits=condition_order
) +
ylab("Novel function gains along lineage") +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
coord_flip() +
facet_wrap(
~extra_task_value,
labeller=label_both
) +
theme(
legend.position="none"
)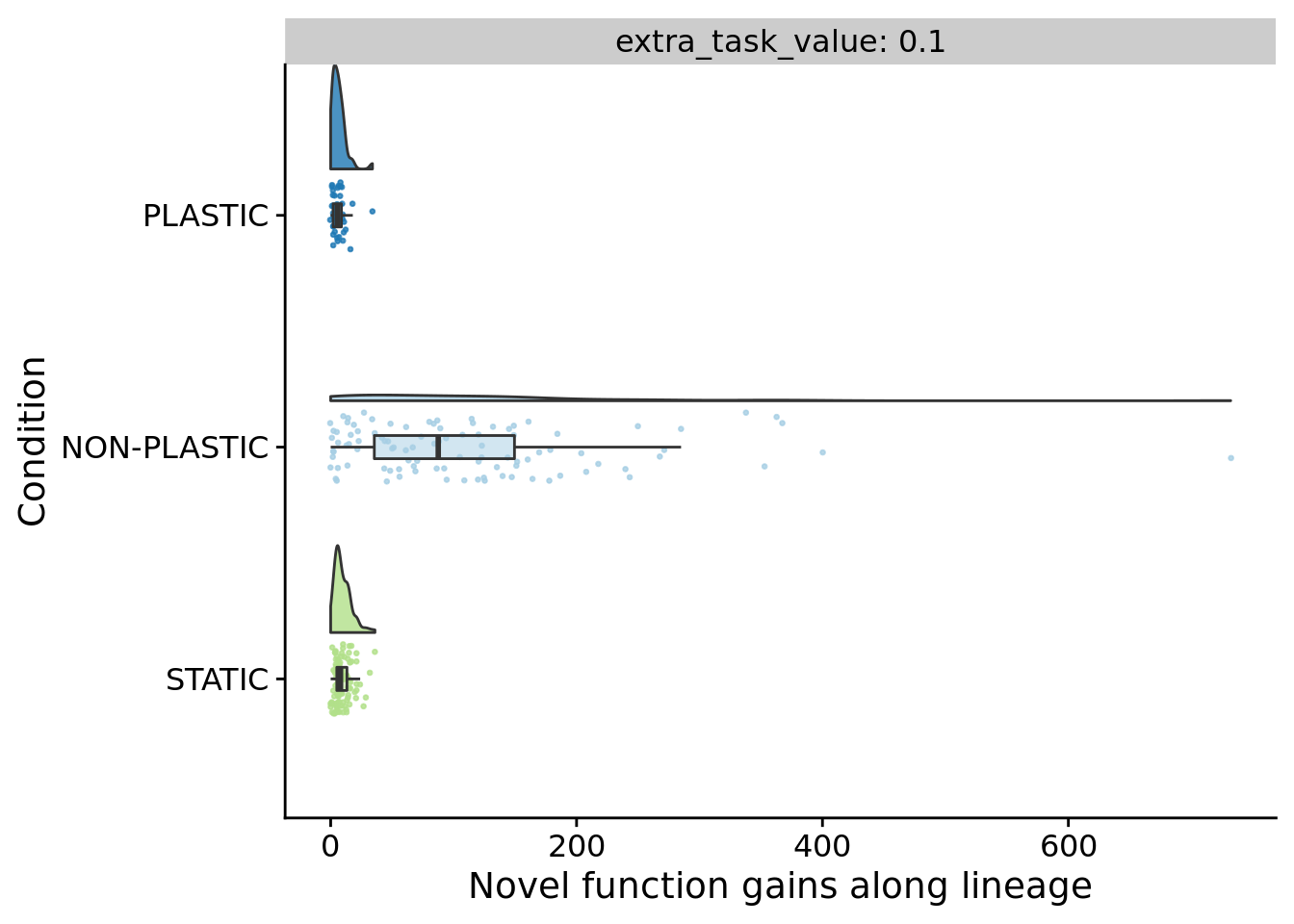
7.11 Novel function loss (lineage)
# Compute manual labels for geom_signif
stat.test <- summary_data %>%
wilcox_test(dominant_lineage_extra_traits_lost ~ condition) %>%
adjust_pvalue(method = "bonferroni") %>%
add_significance() %>%
add_xy_position(x="condition", step.increase=1)
# Tweak y.position manually to account for scaled axis (edge case that triggers bad behavior in geom_signif)
stat.test$manual_position <- log10(stat.test$y.position) * c(1.0,1.0,1.03)
stat.test$label <- mapply(p_label,stat.test$p.adj)
summary_data$is_outlier <- mapply(
is_outlier,
summary_data$dominant_lineage_extra_traits_lost,
summary_data$condition,
MoreArgs=list(data=summary_data, column="dominant_lineage_extra_traits_lost")
)
lineage_novel_task_loss_fig <- ggplot(
summary_data,
aes(x=condition, y=dominant_lineage_extra_traits_lost, fill=condition)
) +
geom_flat_violin(
# data=filter(summary_data,is_outlier==FALSE),
scale="width",
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Condition",
limits=condition_order,
labels=condition_order
) +
scale_y_continuous(
name="Novel function loss (log scale)",
trans=pseudo_log_trans(sigma=1, base=10),
breaks=c(0,10,100,1000),
limits=c(-1,5000)
) +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
labs(
subtitle=paste0(
"Kruskal-Wallis, ",
p_label(signif(kruskal.test(formula=dominant_lineage_extra_traits_lost~condition, data=summary_data)$p.value,digits=4))
)
) +
ggsignif::geom_signif(
data=filter(stat.test, p.adj<=alpha),
aes(xmin=group1,xmax=group2,annotations=label,y_position=manual_position),
manual=TRUE,
inherit.aes=FALSE
) +
# coord_flip()
theme(
legend.position="none"
)## Warning: Ignoring unknown aesthetics: xmin, xmax, annotations, y_position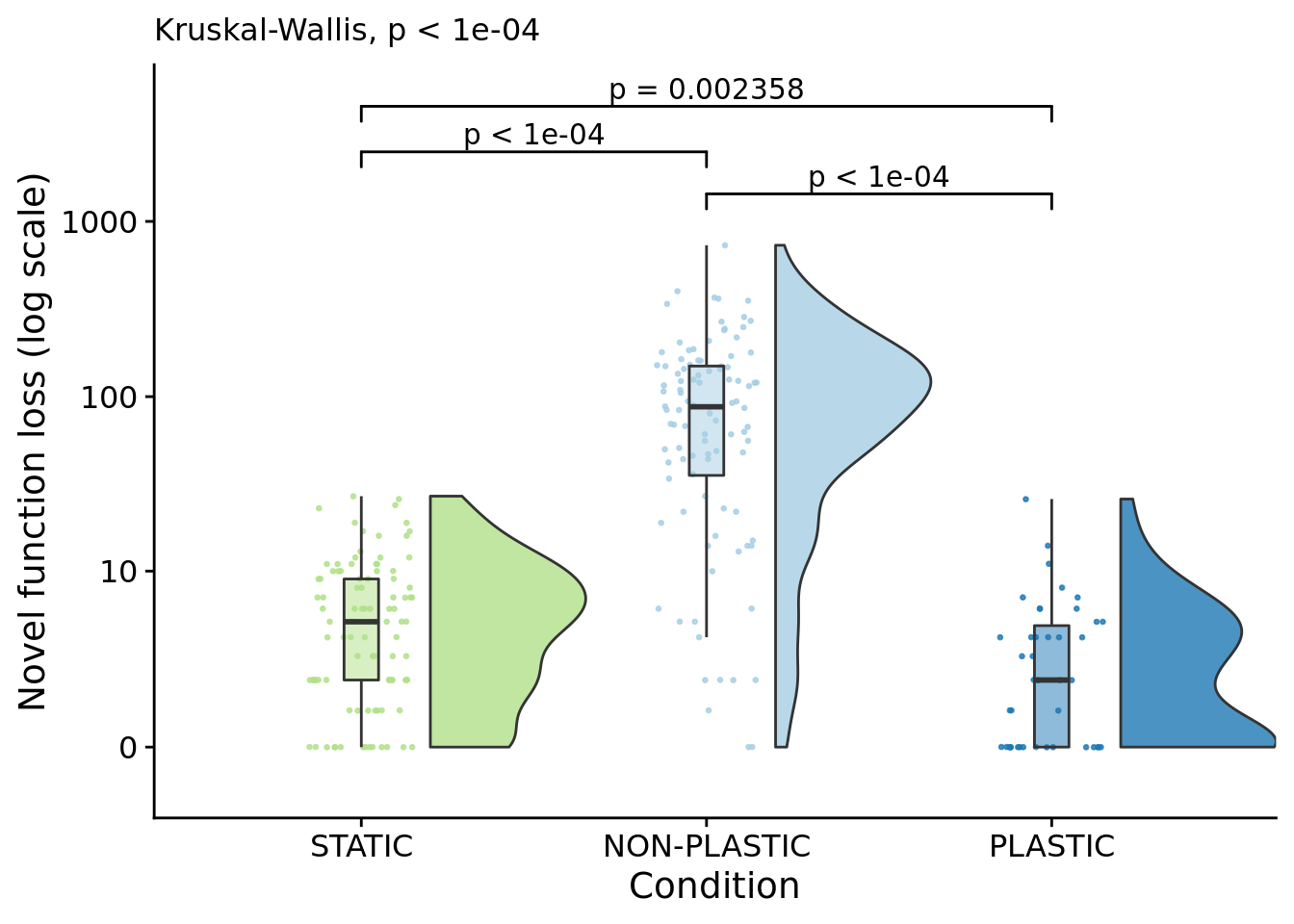
##
## Kruskal-Wallis rank sum test
##
## data: dominant_lineage_extra_traits_lost by condition
## Kruskal-Wallis chi-squared = 129.06, df = 2, p-value < 2.2e-16pairwise.wilcox.test(
x=summary_data$dominant_lineage_extra_traits_lost,
g=summary_data$condition,
p.adjust.method="bonferroni",
conf.int=TRUE,
conf.level=0.95
)##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: summary_data$dominant_lineage_extra_traits_lost and summary_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC 2.7e-16 -
## STATIC < 2e-16 0.0024
##
## P value adjustment method: bonferronipaste(
sep="; ",
paste0(
"PLASTIC median: ",
median(filter(summary_data, condition=="PLASTIC")$dominant_lineage_extra_traits_lost)
),
paste0(
"STATIC median: ",
median(filter(summary_data, condition=="STATIC")$dominant_lineage_extra_traits_lost)
),
paste0(
"NON-PLASTIC median: ",
median(filter(summary_data, condition=="NON-PLASTIC")$dominant_lineage_extra_traits_lost)
)
)## [1] "PLASTIC median: 2; STATIC median: 5; NON-PLASTIC median: 87.5"## [1] "Wilcox rank sum test statistics:"for (pair in pairwise_comparisons) {
pair_data <- filter(summary_data, condition %in% pair)
pair_data$condition <- as.factor(pair_data$condition)
wt <- wilcox.test(
formula=dominant_lineage_extra_traits_lost~condition,
data=pair_data,
exact=FALSE,
paired=FALSE
)
print(paste0(pair[1], "<-->", pair[2], ": W=",wt$statistic))
}## [1] "STATIC<-->NON-PLASTIC: W=9105"
## [1] "STATIC<-->PLASTIC: W=1353.5"
## [1] "PLASTIC<-->NON-PLASTIC: W=3959"7.12 Frequency of novel function loss (lineage)
summary_data$dominant_lineage_extra_traits_lost_per_generation <- summary_data$dominant_lineage_extra_traits_lost / summary_data$dominant_generation_born
summary_data$dominant_lineage_extra_traits_generations_per_loss <- summary_data$dominant_generation_born / summary_data$dominant_lineage_extra_traits_lost
# Compute manual labels for geom_signif
stat.test <- summary_data %>%
wilcox_test(dominant_lineage_extra_traits_lost_per_generation ~ condition) %>%
adjust_pvalue(method = "bonferroni") %>%
add_significance() %>%
add_xy_position(x="condition", step.increase=.1)
# Tweak y.position manually to account for scaled axis (edge case that triggers bad behavior in geom_signif)
stat.test$manual_position <- stat.test$y.position #* c(1.0,1.0,1.03)
stat.test$label <- mapply(p_label,stat.test$p.adj)
summary_data$is_outlier <- mapply(
is_outlier,
summary_data$dominant_lineage_extra_traits_lost_per_generation,
summary_data$condition,
MoreArgs=list(data=summary_data, column="dominant_lineage_extra_traits_lost_per_generation")
)
lineage_novel_task_loss_freq_fig <- ggplot(
# filter(summary_data, dominant_lineage_extra_traits_lost > 0),
summary_data,
aes(x=condition, y=dominant_lineage_extra_traits_lost_per_generation, fill=condition)
) +
geom_flat_violin(
# data=filter(summary_data,is_outlier==FALSE),
scale="width",
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Condition",
limits=condition_order
) +
ylab("Novel function loss frequency") +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
labs(
subtitle=paste0(
"Kruskal-Wallis, ",
p_label(signif(kruskal.test(formula=dominant_lineage_extra_traits_lost_per_generation~condition, data=summary_data)$p.value,digits=4))
)
) +
ggsignif::geom_signif(
data=filter(stat.test, p.adj<=alpha),
aes(xmin=group1,xmax=group2,annotations=label,y_position=manual_position),
manual=TRUE,
inherit.aes=FALSE
) +
theme(
legend.position="none"
)## Warning: Ignoring unknown aesthetics: xmin, xmax, annotations, y_position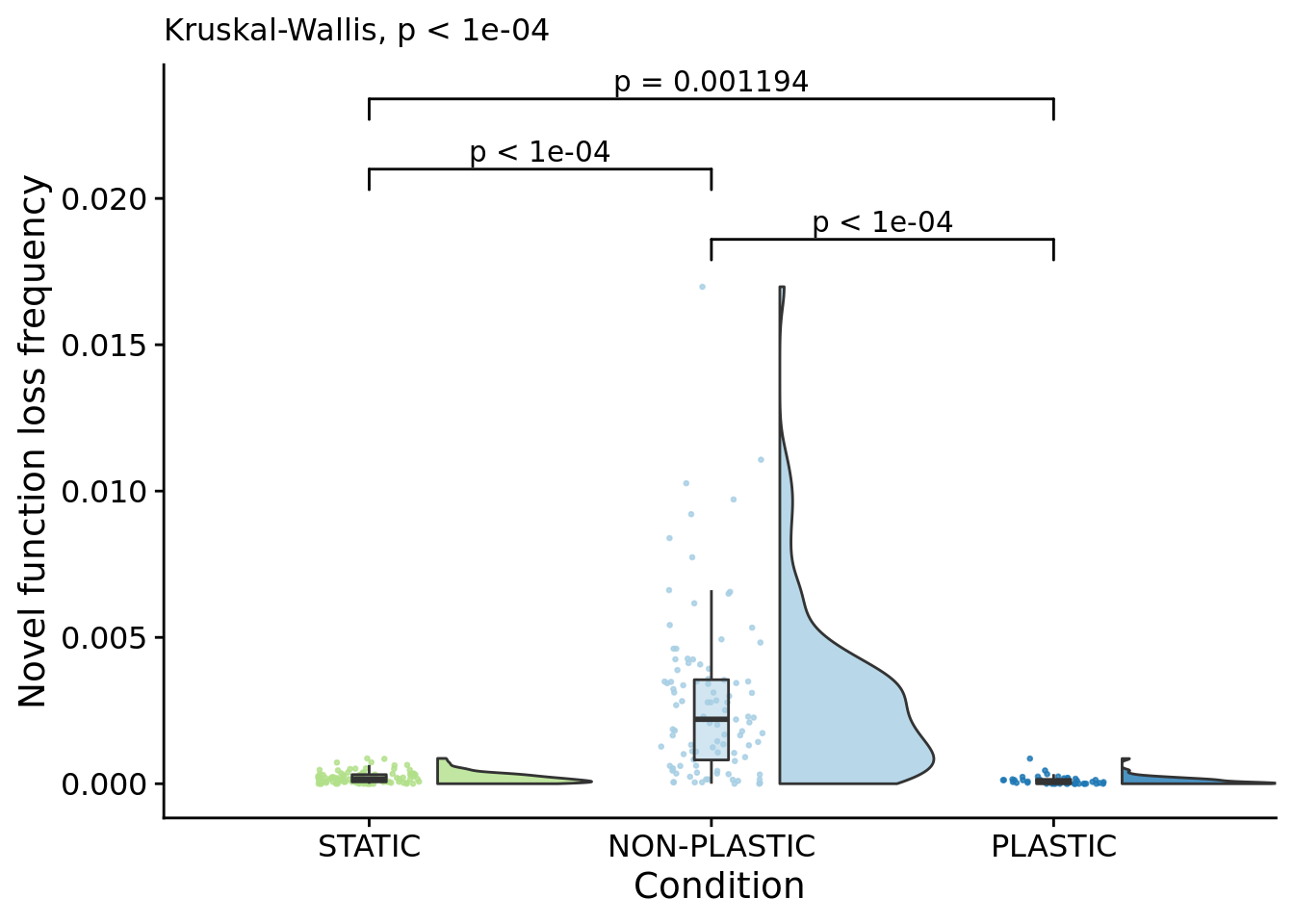
kruskal.test(
formula=dominant_lineage_extra_traits_lost_per_generation~condition,
data=summary_data
)##
## Kruskal-Wallis rank sum test
##
## data: dominant_lineage_extra_traits_lost_per_generation by condition
## Kruskal-Wallis chi-squared = 121.41, df = 2, p-value < 2.2e-16pairwise.wilcox.test(
x=summary_data$dominant_lineage_extra_traits_lost_per_generation,
g=summary_data$condition,
p.adjust.method="bonferroni",
conf.int=TRUE,
conf.level=0.95
)##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: summary_data$dominant_lineage_extra_traits_lost_per_generation and summary_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC 1.1e-15 -
## STATIC < 2e-16 0.0012
##
## P value adjustment method: bonferronipaste(
sep="; ",
paste0(
"PLASTIC median: ",
median(filter(summary_data, condition=="PLASTIC")$dominant_lineage_extra_traits_lost_per_generation)
),
paste0(
"STATIC median: ",
median(filter(summary_data, condition=="STATIC")$dominant_lineage_extra_traits_lost_per_generation)
),
paste0(
"NON-PLASTIC median: ",
median(filter(summary_data, condition=="NON-PLASTIC")$dominant_lineage_extra_traits_lost_per_generation)
)
)## [1] "PLASTIC median: 6.25141973661864e-05; STATIC median: 0.000161396283669756; NON-PLASTIC median: 0.0022026054610079"## [1] "Wilcox rank sum test statistics:"for (pair in pairwise_comparisons) {
pair_data <- filter(summary_data, condition %in% pair)
pair_data$condition <- as.factor(pair_data$condition)
wt <- wilcox.test(
formula=dominant_lineage_extra_traits_lost_per_generation~condition,
data=pair_data,
exact=FALSE,
paired=FALSE
)
print(paste0(pair[1], "<-->", pair[2], ": W=",wt$statistic))
}## [1] "STATIC<-->NON-PLASTIC: W=8940"
## [1] "STATIC<-->PLASTIC: W=1311"
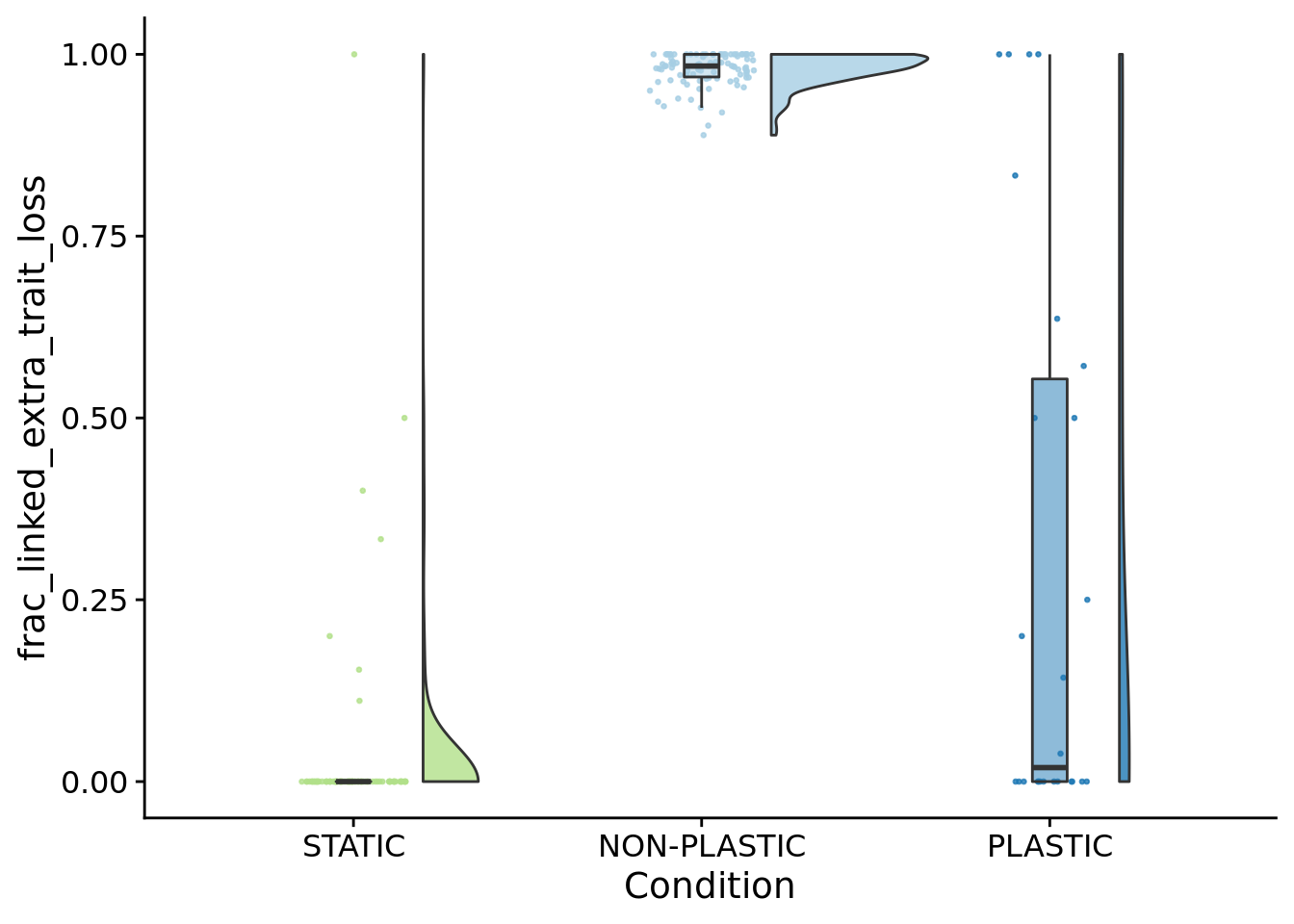
## [1] "PLASTIC<-->NON-PLASTIC: W=3922"7.13 How many instances of novel function loss co-occurred with changes in base phenotype?
Function loss linked with base function changes.
lost_traits_summary_data <- filter(summary_data, extra_task_value==0.1 & dominant_lineage_extra_traits_lost>0)
lost_traits_summary_data$frac_linked_extra_trait_loss <- lost_traits_summary_data$dominant_lineage_extra_traits_lost_linked_to_primary_change / lost_traits_summary_data$dominant_lineage_extra_traits_lostggplot(lost_traits_summary_data, aes(x=condition, y=frac_linked_extra_trait_loss, fill=condition)) +
geom_flat_violin(
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Condition",
limits=condition_order
) +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
theme(
legend.position="none"
)
##
## Kruskal-Wallis rank sum test
##
## data: frac_linked_extra_trait_loss by condition
## Kruskal-Wallis chi-squared = 153.68, df = 2, p-value < 2.2e-16pairwise.wilcox.test(
x=lost_traits_summary_data$frac_linked_extra_trait_loss,
g=lost_traits_summary_data$condition,
p.adjust.method="bonferroni",
conf.int=TRUE,
conf.level=0.95
)##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: lost_traits_summary_data$frac_linked_extra_trait_loss and lost_traits_summary_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC 1.9e-08 -
## STATIC < 2e-16 1.8e-06
##
## P value adjustment method: bonferronipaste(
sep="; ",
paste0(
"PLASTIC median: ",
median(filter(lost_traits_summary_data, condition=="PLASTIC")$frac_linked_extra_trait_loss)
),
paste0(
"STATIC median: ",
median(filter(lost_traits_summary_data, condition=="STATIC")$frac_linked_extra_trait_loss)
),
paste0(
"NON-PLASTIC median: ",
median(filter(lost_traits_summary_data, condition=="NON-PLASTIC")$frac_linked_extra_trait_loss)
)
)## [1] "PLASTIC median: 0.0192307692307692; STATIC median: 0; NON-PLASTIC median: 0.983803278688525"## [1] "Wilcox rank sum test statistics:"for (pair in pairwise_comparisons) {
pair_data <- filter(lost_traits_summary_data, condition %in% pair)
pair_data$condition <- as.factor(pair_data$condition)
wt <- wilcox.test(
formula=frac_linked_extra_trait_loss~condition,
data=pair_data,
exact=FALSE,
paired=FALSE
)
print(paste0(pair[1], "<-->", pair[2], ": W=",wt$statistic))
}## [1] "STATIC<-->NON-PLASTIC: W=8344"
## [1] "STATIC<-->PLASTIC: W=1602"
## [1] "PLASTIC<-->NON-PLASTIC: W=2212"## [1] 10998## [1] 11229aggregate_frac_linked_extra_trait_loss_nonplastic <- sum(filter(lost_traits_summary_data, condition=="NON-PLASTIC")$dominant_lineage_extra_traits_lost_linked_to_primary_change) / sum(filter(lost_traits_summary_data, condition=="NON-PLASTIC")$dominant_lineage_extra_traits_lost)
aggregate_frac_linked_extra_trait_loss_nonplastic## [1] 0.9794283## [1] 29## [1] 142aggregate_frac_linked_extra_trait_loss_plastic <- sum(filter(lost_traits_summary_data, condition=="PLASTIC")$dominant_lineage_extra_traits_lost_linked_to_primary_change) / sum(filter(lost_traits_summary_data, condition=="PLASTIC")$dominant_lineage_extra_traits_lost)
aggregate_frac_linked_extra_trait_loss_plastic## [1] 0.2042254## [1] 13## [1] 631aggregate_frac_linked_extra_trait_loss_nonplastic <- sum(filter(lost_traits_summary_data, condition=="STATIC")$dominant_lineage_extra_traits_lost_linked_to_primary_change) / sum(filter(lost_traits_summary_data, condition=="STATIC")$dominant_lineage_extra_traits_lost)
aggregate_frac_linked_extra_trait_loss_nonplastic## [1] 0.020602227.15 Combined panel
magnitude_grid <- plot_grid(
final_novel_task_count_fig +
theme(
axis.title.x=element_blank()
) +
ggtitle("Final novel function count"),
lineage_novel_task_discovery_fig +
theme(
axis.title.x=element_blank()
) +
ggtitle("Novel function discovery"),
lineage_novel_task_loss_fig +
theme(
axis.title.x=element_blank()
) +
ggtitle("Novel function loss"),
nrow=1,
align="v",
labels="auto"
)
magnitude_grid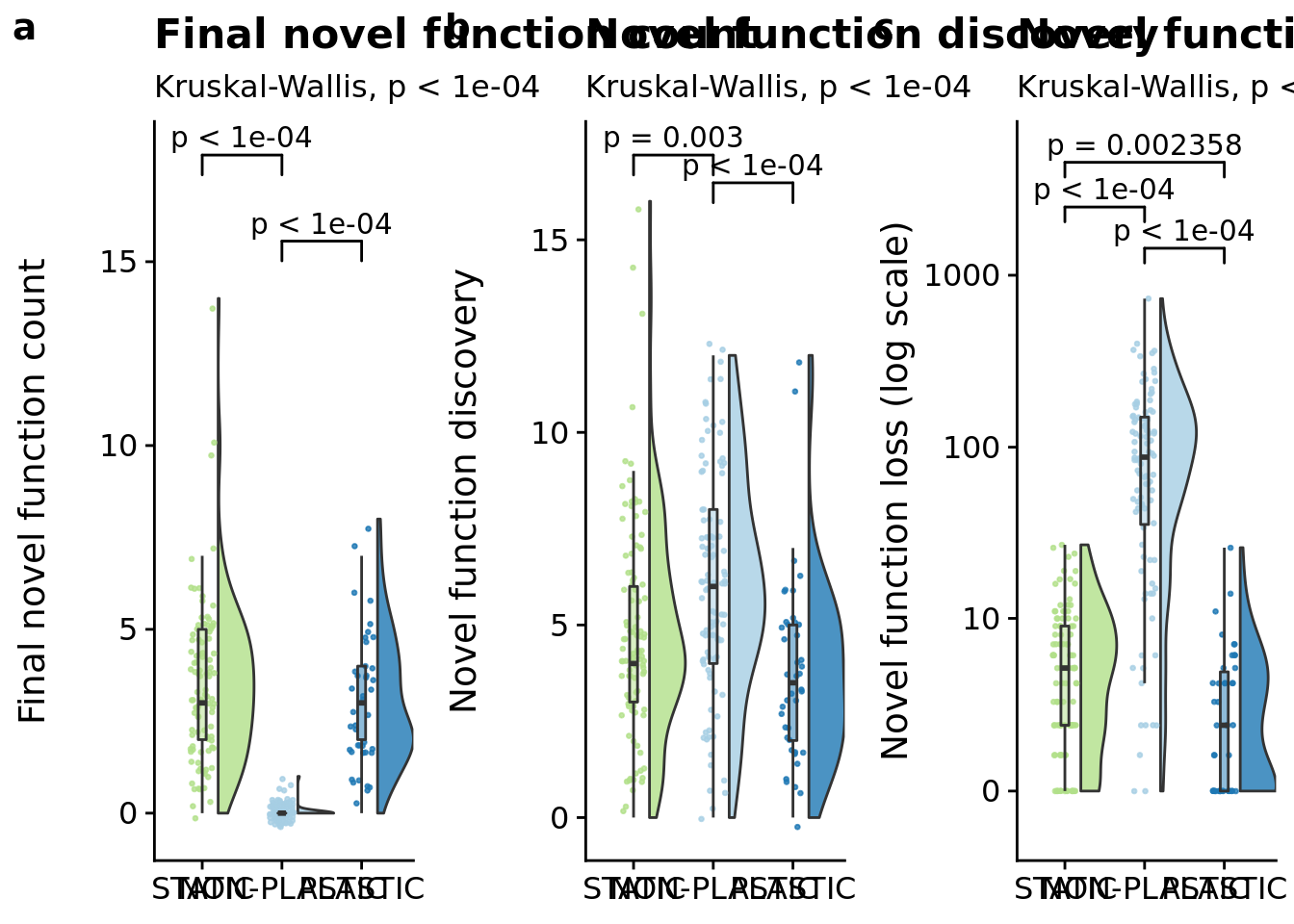
pace_grid <- plot_grid(
lineage_novel_task_discovery_freq_fig +
theme(
axis.title.x=element_blank()
) +
ggtitle("Novel function discovery frequency"),
lineage_novel_task_loss_freq_fig +
theme(
axis.title.x=element_blank()
) +
ggtitle("Novel function loss frequency"),
nrow=1,
align="v",
labels="auto"
)
pace_grid