Chapter 6 Evolutionary change
The effect of adaptive phenotypic plasticity on evolutionary change.
6.1 Overview
total_updates <- 200000
replicates <- 100
alpha <- 0.05
all_traits <- c("not","nand","and","ornot","or","andnot")
traits_set_a <- c("not", "and", "or")
traits_set_b <- c("nand", "ornot", "andnot")
# Relative location of data.
working_directory <- "experiments/2021-02-08-evo-dynamics/analysis/" # << For bookdown
# working_directory <- "./" # << For local analysis6.2 Analysis dependencies
Load all required R libraries.
library(ggplot2)
library(rstatix)
library(ggsignif)
library(scales)
library(tidyverse)
library(cowplot)
library(RColorBrewer)
library(Hmisc)
library(boot)
source("https://gist.githubusercontent.com/benmarwick/2a1bb0133ff568cbe28d/raw/fb53bd97121f7f9ce947837ef1a4c65a73bffb3f/geom_flat_violin.R")These analyses were conducted/knitted with the following computing environment:
## _
## platform x86_64-pc-linux-gnu
## arch x86_64
## os linux-gnu
## system x86_64, linux-gnu
## status
## major 4
## minor 1.3
## year 2022
## month 03
## day 10
## svn rev 81868
## language R
## version.string R version 4.1.3 (2022-03-10)
## nickname One Push-Up6.3 Setup
summary_data_loc <- paste0(working_directory, "data/aggregate.csv")
summary_data <- read.csv(summary_data_loc, na.strings="NONE")
summary_data$DISABLE_REACTION_SENSORS <- as.factor(summary_data$DISABLE_REACTION_SENSORS)
summary_data$chg_env <- summary_data$chg_env == "True"
summary_data$dominant_plastic_odd_even <- as.factor(summary_data$dominant_plastic_odd_even)
summary_data$sensors <- summary_data$DISABLE_REACTION_SENSORS == "0"
summary_data$is_plastic <- summary_data$dominant_plastic_odd_even == "True"
env_label_fun <- function(chg_env) {
if (chg_env) {
return("Fluctuating")
} else {
return("Constant")
}
}
sensors_label_fun <- function(has_sensors) {
if (has_sensors) {
return("Sensors")
} else {
return("No sensors")
}
}
# note that this labeler makes assumptions about how we set up our experiment
condition_label_fun <- function(has_sensors, env_chg) {
if (has_sensors && env_chg) {
return("PLASTIC")
} else if (env_chg) {
return("NON-PLASTIC")
} else {
return("STATIC")
}
}
summary_data$env_label <- mapply(
env_label_fun,
summary_data$chg_env
)
summary_data$sensors_label <- mapply(
sensors_label_fun,
summary_data$sensors
)
summary_data$condition <- mapply(
condition_label_fun,
summary_data$sensors,
summary_data$chg_env
)
condition_order = c(
"STATIC",
"NON-PLASTIC",
"PLASTIC"
)
pairwise_comparisons <- list(
c("STATIC", "NON-PLASTIC"),
c("STATIC", "PLASTIC"),
c("PLASTIC", "NON-PLASTIC")
)
p_label <- function(p_value) {
threshold = 0.0001
if (p_value < threshold) {
return(paste0("p < ", threshold))
} else {
return(paste0("p = ", p_value))
}
}
# *really* inefficient way to identify outliers
is_outlier <- function(value, cond, data, column) {
cond_data <- filter(data, condition==cond)
q1 <- summary(cond_data[,column])[["1st Qu."]]
q3 <- summary(cond_data[,column])[["3rd Qu."]]
H <- 1.5 * IQR(cond_data[,column])
return( (value < (q1-H)) || (value > (q3+H)) )
}
####### misc #######
# Configure our default graphing theme
theme_set(theme_cowplot())
# Palette
cb_palette <- "Paired"
# Create a directory to store plots
dir.create(paste0(working_directory, "plots"), showWarnings=FALSE)
# Define sample mean function
samplemean <- function(x, d) {
return(mean(x[d]))
}6.4 The evolution of phenotypic plasticity
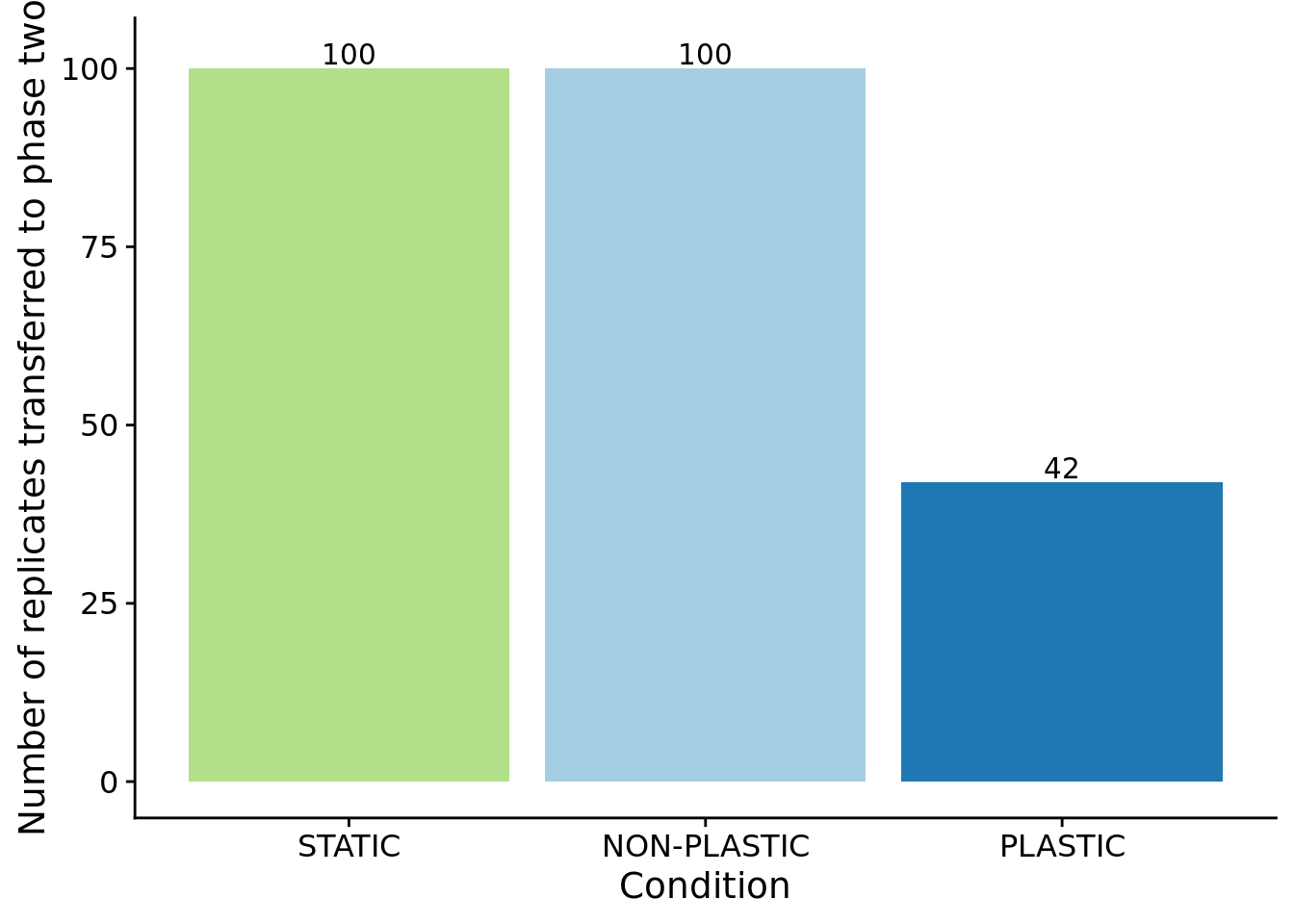
For sensor-enabled populations in fluctuating environments, we only transfered populations containing an optimally plastic genotype to phase-two.
summary_data_grouped = dplyr::group_by(summary_data, condition)
summary_data_group_counts = dplyr::summarize(summary_data_grouped, n=dplyr::n())
ggplot(summary_data_group_counts, aes(x=condition, y=n, fill=condition)) +
geom_col(position=position_dodge(0.9)) +
geom_text(aes(label=n, y=n+2)) +
scale_x_discrete(
name="Condition",
limits=condition_order
) +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
ylab("Number of replicates transferred to phase two") +
theme(
legend.position="none"
)
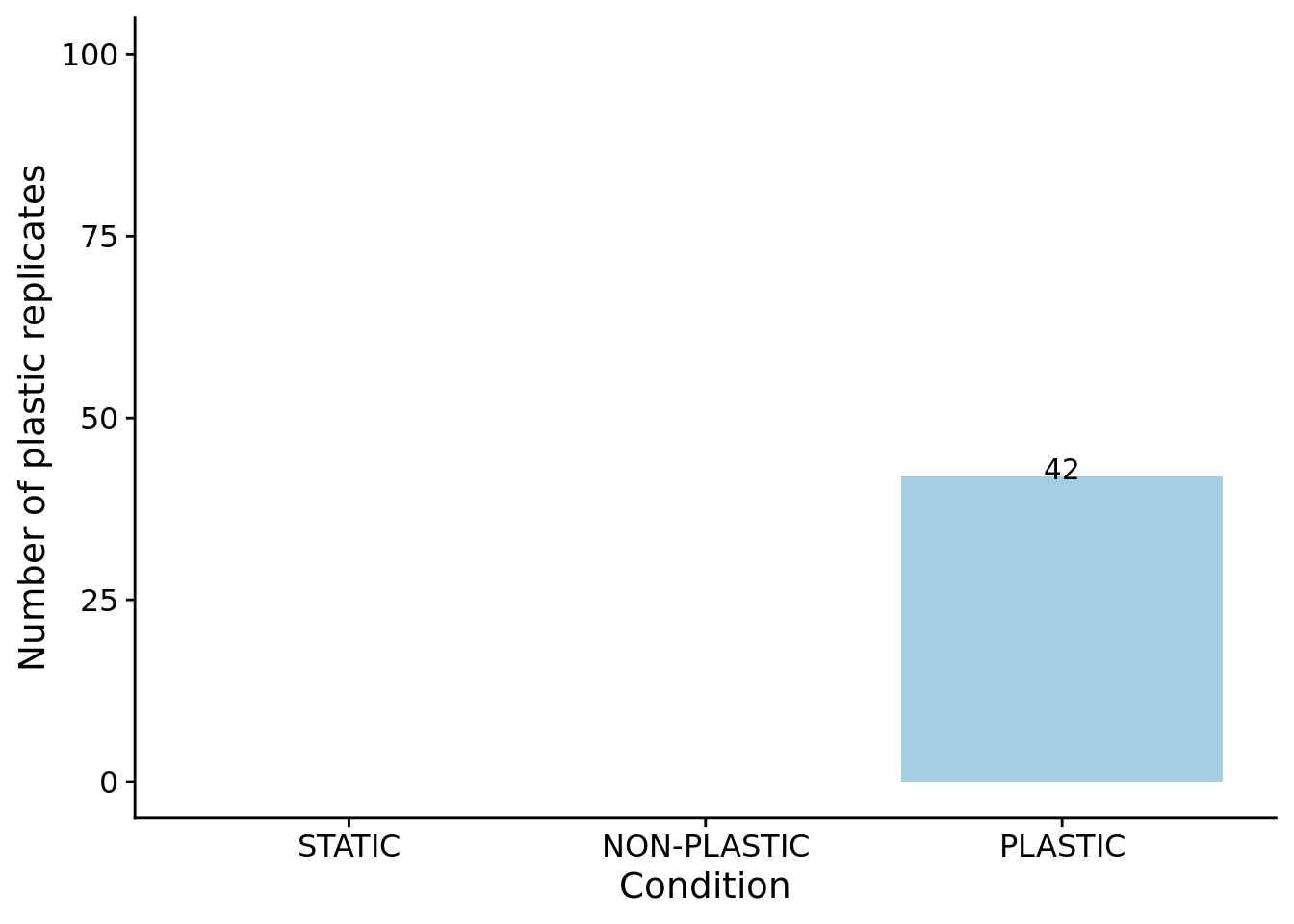
We can confirm our expectation that the dominant genotypes in non-plastic conditions are not phenotypically plastic.
summary_data_grouped = dplyr::group_by(summary_data, condition, is_plastic)
summary_data_group_counts = dplyr::summarize(summary_data_grouped, n=dplyr::n())
ggplot(filter(summary_data_group_counts, is_plastic), aes(x=condition, y=n, fill=condition)) +
geom_col(
position=position_dodge(0.9)
) +
scale_x_discrete(
name="Condition",
limits=condition_order
) +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
geom_text(aes(label=n, y=n+1)) +
ylab("Number of plastic replicates") +
ylim(0, 100) +
theme(
legend.position="none"
)
6.5 Average generation
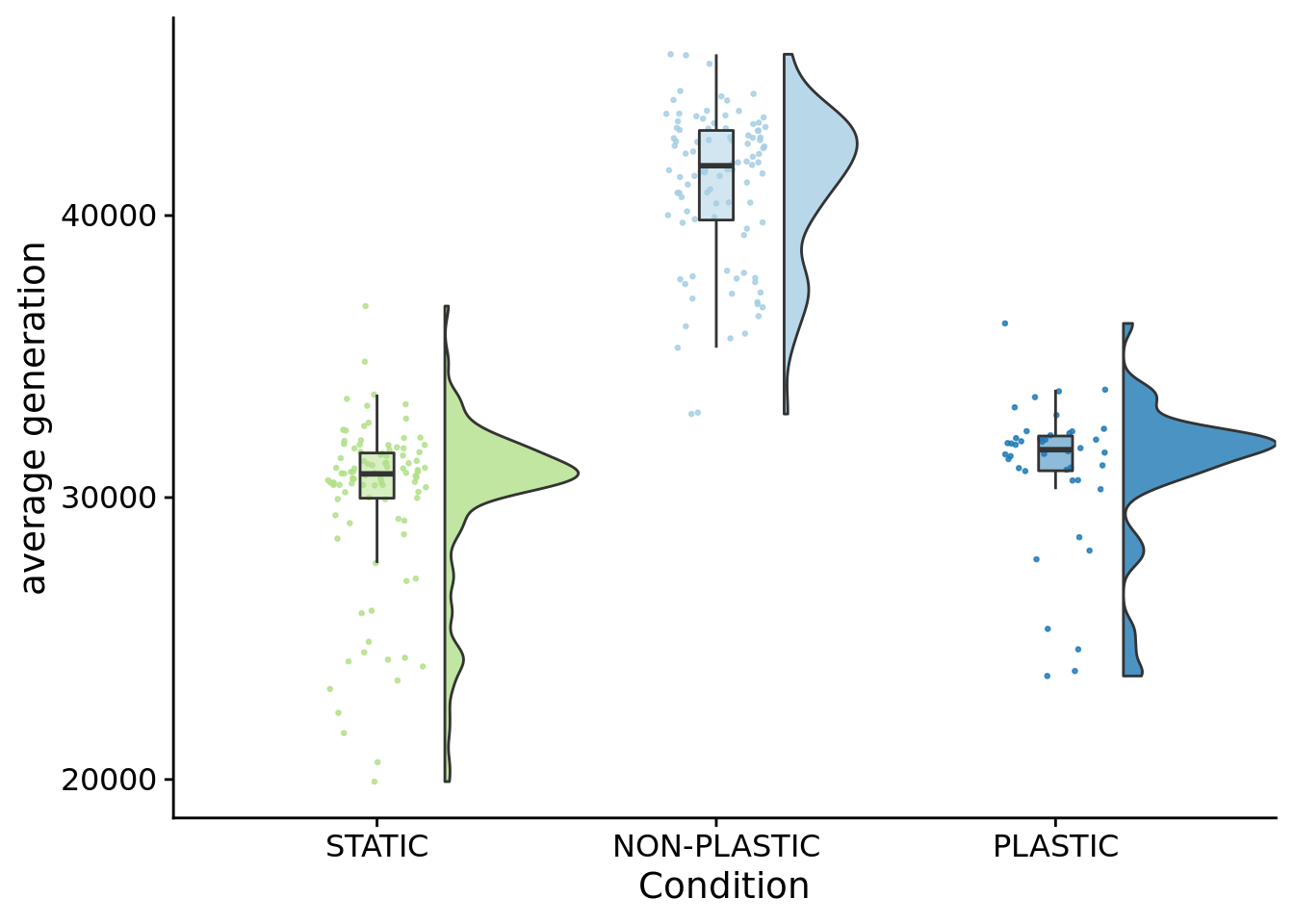
How many generations elapsed in each of of our treatments?
ggplot(summary_data, aes(x=condition, y=time_average_generation, fill=condition)) +
geom_flat_violin(
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Condition",
limits=condition_order
) +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
# coord_flip() +
ylab("average generation") +
theme(
legend.position="none"
)
## Saving 7 x 5 in image##
## Kruskal-Wallis rank sum test
##
## data: time_average_generation by condition
## Kruskal-Wallis chi-squared = 177.33, df = 2, p-value < 2.2e-16pairwise.wilcox.test(
x=summary_data$time_average_generation,
g=summary_data$condition,
p.adjust.method="bonferroni",
)##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: summary_data$time_average_generation and summary_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC <2e-16 -
## STATIC <2e-16 0.004
##
## P value adjustment method: bonferronipaste(
sep="; ",
paste0(
"PLASTIC median: ",
median(filter(summary_data, condition=="PLASTIC")$time_average_generation)
),
paste0(
"STATIC median: ",
median(filter(summary_data, condition=="STATIC")$time_average_generation)
),
paste0(
"NON-PLASTIC median: ",
median(filter(summary_data, condition=="NON-PLASTIC")$time_average_generation)
)
)## [1] "PLASTIC median: 31697.65; STATIC median: 30839.75; NON-PLASTIC median: 41768.65"## [1] "Wilcox rank sum test statistics:"for (pair in pairwise_comparisons) {
pair_data <- filter(summary_data, condition %in% pair)
pair_data$condition <- as.factor(pair_data$condition)
wt <- wilcox.test(
formula=time_average_generation~condition,
data=pair_data,
exact=FALSE,
paired=FALSE
)
print(paste0(pair[1], "<-->", pair[2], ": W=",wt$statistic))
}## [1] "STATIC<-->NON-PLASTIC: W=9982"
## [1] "STATIC<-->PLASTIC: W=2818"
## [1] "PLASTIC<-->NON-PLASTIC: W=4186"summary_data %>%
group_by(condition) %>%
summarise(mean=mean(time_average_generation),sd=sd(time_average_generation))## # A tibble: 3 x 3
## condition mean sd
## <chr> <dbl> <dbl>
## 1 NON-PLASTIC 41090. 2702.
## 2 PLASTIC 31016. 2615.
## 3 STATIC 30002. 3011.6.6 Coalescence event count
The number of times the most recent common ancestor changes gives us the number of selective sweeps that occur during the experiment.
# Compute manual labels for geom_signif
stat.test <- summary_data %>%
wilcox_test(phylo_mrca_changes ~ condition) %>%
adjust_pvalue(method = "bonferroni") %>%
add_significance() %>%
add_xy_position(x="condition",step.increase=1)
# Tweak y.position manually to account for scaled axis (edge case that triggers bad behavior in geom_signif)
stat.test$manual_position <- log10(stat.test$y.position) * c(1.0,1.0,1.03)
stat.test$label <- mapply(p_label,stat.test$p.adj)
summary_data$is_outlier <- mapply(
is_outlier,
summary_data$phylo_mrca_changes,
summary_data$condition,
MoreArgs=list(data=summary_data, column="phylo_mrca_changes")
)
coalescence_events_fig <- ggplot(
summary_data,
aes(x=condition, y=phylo_mrca_changes,fill=condition)
) +
geom_flat_violin(
# data=filter(summary_data,is_outlier==FALSE),
scale="width",
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Condition",
limits=condition_order,
labels=condition_order,
breaks=condition_order
) +
scale_y_continuous(
name="Coalescence event count (log scale)",
trans=pseudo_log_trans(sigma = 1, base = 10),
breaks=c(0, 10, 100, 1000, 10000),
limits=c(-1, 35000)
) +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
labs(
subtitle=paste0(
"Kruskal-Wallis, ",
p_label(signif(kruskal.test(formula=phylo_mrca_changes~condition, data=summary_data)$p.value,digits=4))
)
) +
ggsignif::geom_signif(
data=filter(stat.test, p.adj <= alpha),
aes(xmin=group1,xmax=group2,annotations=label,y_position=manual_position),
manual=TRUE,
inherit.aes=FALSE
) +
theme(
legend.position="none"
)## Warning: Ignoring unknown aesthetics: xmin, xmax, annotations, y_positionggsave(
paste0(working_directory, "plots/", "selective-sweeps.pdf"),
width=5,
height=5
)
coalescence_events_fig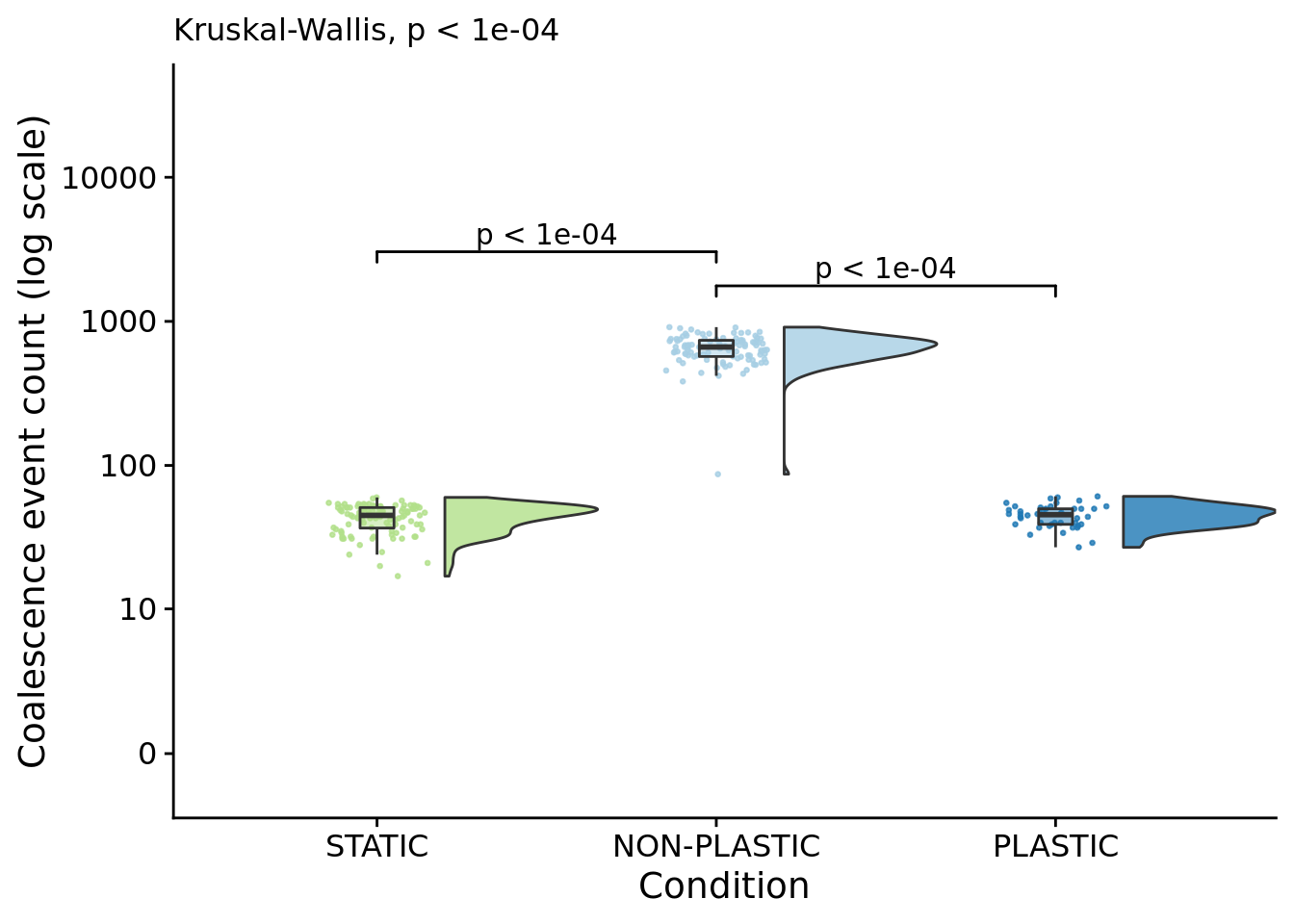
##
## Kruskal-Wallis rank sum test
##
## data: phylo_mrca_changes by condition
## Kruskal-Wallis chi-squared = 175.46, df = 2, p-value < 2.2e-16pairwise.wilcox.test(
x=summary_data$phylo_mrca_changes,
g=summary_data$condition,
p.adjust.method="bonferroni",
)##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: summary_data$phylo_mrca_changes and summary_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC <2e-16 -
## STATIC <2e-16 1
##
## P value adjustment method: bonferronipaste(
sep="; ",
paste0(
"PLASTIC median: ",
median(filter(summary_data, condition=="PLASTIC")$phylo_mrca_changes)
),
paste0(
"STATIC median: ",
median(filter(summary_data, condition=="STATIC")$phylo_mrca_changes)
),
paste0(
"NON-PLASTIC median: ",
median(filter(summary_data, condition=="NON-PLASTIC")$phylo_mrca_changes)
)
)## [1] "PLASTIC median: 45.5; STATIC median: 45; NON-PLASTIC median: 663.5"## [1] "Wilcox rank sum test statistics:"for (pair in pairwise_comparisons) {
pair_data <- filter(summary_data, condition %in% pair)
pair_data$condition <- as.factor(pair_data$condition)
wt <- wilcox.test(
formula=phylo_mrca_changes~condition,
data=pair_data,
exact=FALSE,
paired=FALSE
)
print(paste0(pair[1], "<-->", pair[2], ": W=",wt$statistic))
}## [1] "STATIC<-->NON-PLASTIC: W=10000"
## [1] "STATIC<-->PLASTIC: W=2215"
## [1] "PLASTIC<-->NON-PLASTIC: W=4200"6.6.1 Average number of generations between coalescence events
# Compute frequency of coalescence events
summary_data$generations_per_mrca_change <- summary_data$time_average_generation / summary_data$phylo_mrca_changes
# Compute manual labels for geom_signif
stat.test <- summary_data %>%
wilcox_test(generations_per_mrca_change ~ condition) %>%
adjust_pvalue(method = "bonferroni") %>%
add_significance() %>%
add_xy_position(x="condition")
# Tweak y.position manually to account for scaled axis (edge case that triggers bad behavior in geom_signif)
stat.test$manual_position <- stat.test$y.position
stat.test$label <- mapply(p_label,stat.test$p.adj)
summary_data$is_outlier <- mapply(
is_outlier,
summary_data$generations_per_mrca_change,
summary_data$condition,
MoreArgs=list(data=summary_data, column="generations_per_mrca_change")
)
coalescence_events_freq_fig <- ggplot(
summary_data,
aes(x=condition, y=generations_per_mrca_change, fill=condition)
) +
geom_flat_violin(
# data=filter(summary_data,is_outlier==FALSE),
scale="width",
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Condition",
limits=condition_order,
labels=condition_order
) +
scale_y_continuous(
name="Avg. generations between coalescence events",
limits=c(0, 2000),
breaks=seq(0, 2000, 500)
) +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
# coord_flip() +
labs(
subtitle=paste0(
"Kruskal-Wallis, ",
p_label(signif(kruskal.test(formula=generations_per_mrca_change~condition, data=summary_data)$p.value,digits=4))
)
) +
ggsignif::geom_signif(
data=filter(stat.test, p.adj <= alpha),
aes(xmin=group1,xmax=group2,annotations=label,y_position=manual_position),
manual=TRUE,
inherit.aes=FALSE
) +
theme(
legend.position="none"
)## Warning: Ignoring unknown aesthetics: xmin, xmax, annotations, y_positionggsave(
paste0(working_directory, "plots/", "generations-between-selective-sweeps.png"),
width=5,
height=5
)
coalescence_events_freq_fig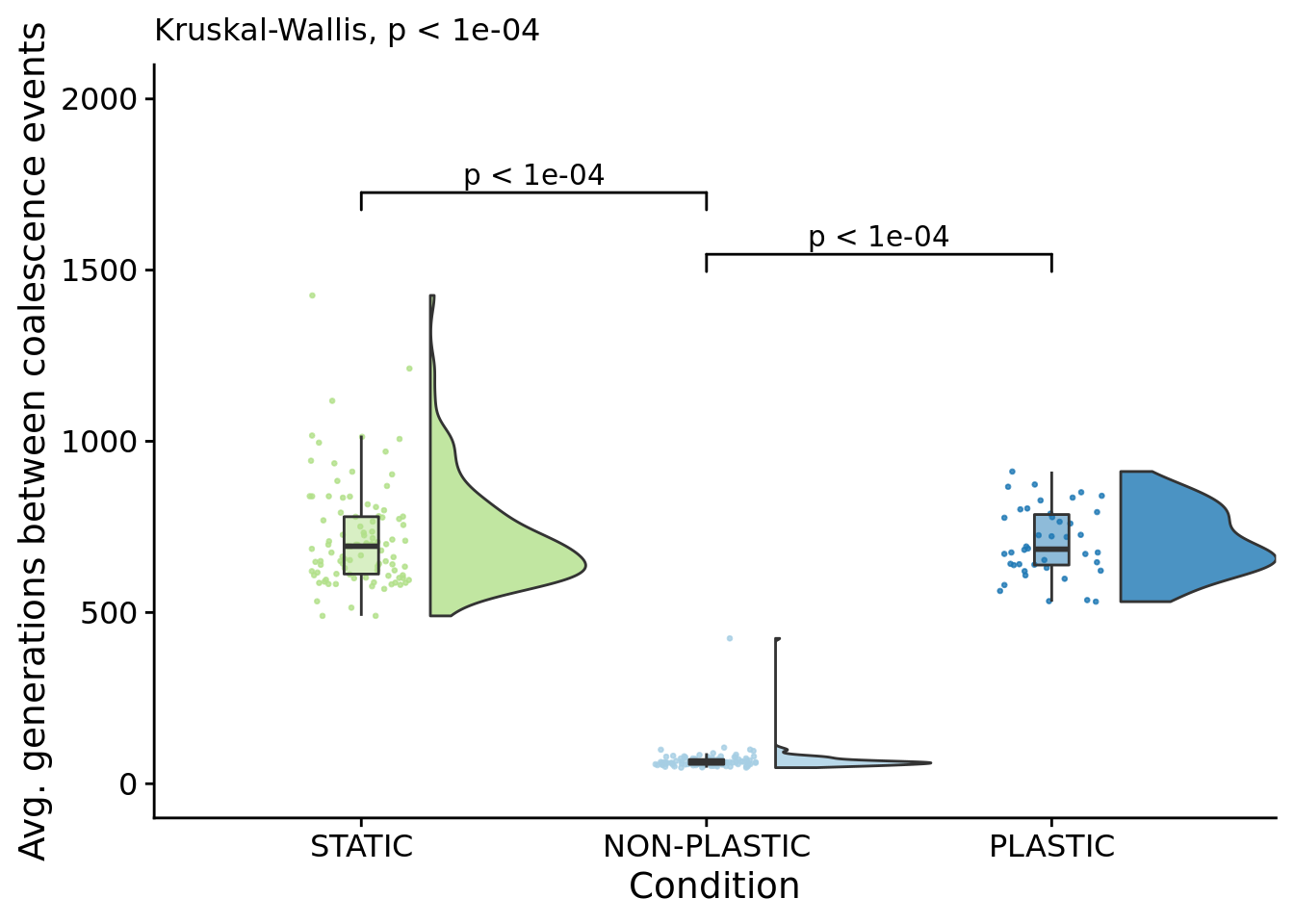
##
## Kruskal-Wallis rank sum test
##
## data: generations_per_mrca_change by condition
## Kruskal-Wallis chi-squared = 175.33, df = 2, p-value < 2.2e-16pairwise.wilcox.test(
x=summary_data$generations_per_mrca_change,
g=summary_data$condition,
p.adjust.method="bonferroni",
)##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: summary_data$generations_per_mrca_change and summary_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC <2e-16 -
## STATIC <2e-16 1
##
## P value adjustment method: bonferronipaste(
sep="; ",
paste0(
"PLASTIC median: ",
median(filter(summary_data, condition=="PLASTIC")$generations_per_mrca_change)
),
paste0(
"STATIC median: ",
median(filter(summary_data, condition=="STATIC")$generations_per_mrca_change)
),
paste0(
"NON-PLASTIC median: ",
median(filter(summary_data, condition=="NON-PLASTIC")$generations_per_mrca_change)
)
)## [1] "PLASTIC median: 685.001780758557; STATIC median: 693.676265008576; NON-PLASTIC median: 62.0184902295191"## [1] "Wilcox rank sum test statistics:"for (pair in pairwise_comparisons) {
pair_data <- filter(summary_data, condition %in% pair)
pair_data$condition <- as.factor(pair_data$condition)
wt <- wilcox.test(
formula=generations_per_mrca_change~condition,
data=pair_data,
exact=FALSE,
paired=FALSE
)
print(paste0(pair[1], "<-->", pair[2], ": W=",wt$statistic))
}## [1] "STATIC<-->NON-PLASTIC: W=0"
## [1] "STATIC<-->PLASTIC: W=2151"
## [1] "PLASTIC<-->NON-PLASTIC: W=0"6.7 Phenotypic volatility along the dominant lineage
# Compute manual labels for geom_signif
stat.test <- summary_data %>%
wilcox_test(dominant_lineage_trait_volatility ~ condition) %>%
adjust_pvalue(method = "bonferroni") %>%
add_significance() %>%
add_xy_position(x="condition", step.increase=1)
# Tweak y.position manually to account for scaled axis (edge case that triggers bad behavior in geom_signif)
stat.test$manual_position <- log10(stat.test$y.position) * c(1.0,1.0,1.03)
stat.test$label <- mapply(p_label,stat.test$p.adj)
summary_data$is_outlier <- mapply(
is_outlier,
summary_data$dominant_lineage_trait_volatility,
summary_data$condition,
MoreArgs=list(data=summary_data, column="dominant_lineage_trait_volatility")
)
phenotypic_volatility_fig <- ggplot(
summary_data,
aes(x=condition, y=dominant_lineage_trait_volatility, fill=condition)
) +
geom_flat_violin(
# data=filter(summary_data,is_outlier==FALSE),
scale="width",
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Condition",
limits=condition_order,
labels=condition_order
) +
scale_y_continuous(
name="Phenotypic volatility (log scale)",
trans=pseudo_log_trans(sigma = 1, base = 10),
breaks=c(0, 10, 100, 1000, 10000),
limits=c(-1, 35000)
) +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
labs(
subtitle=paste0(
"Kruskal-Wallis, ",
p_label(signif(kruskal.test(formula=dominant_lineage_trait_volatility~condition, data=summary_data)$p.value,digits=4))
)
) +
ggsignif::geom_signif(
data=filter(stat.test, p.adj<=alpha),
aes(xmin=group1,xmax=group2,annotations=label,y_position=manual_position),
manual=TRUE,
inherit.aes=FALSE
) +
# coord_flip() +
theme(
legend.position="none"
)## Warning: Ignoring unknown aesthetics: xmin, xmax, annotations, y_positionggsave(
paste0(working_directory, "plots/", "phenotypic-volatility.pdf"),
width=5,
height=5
)
phenotypic_volatility_fig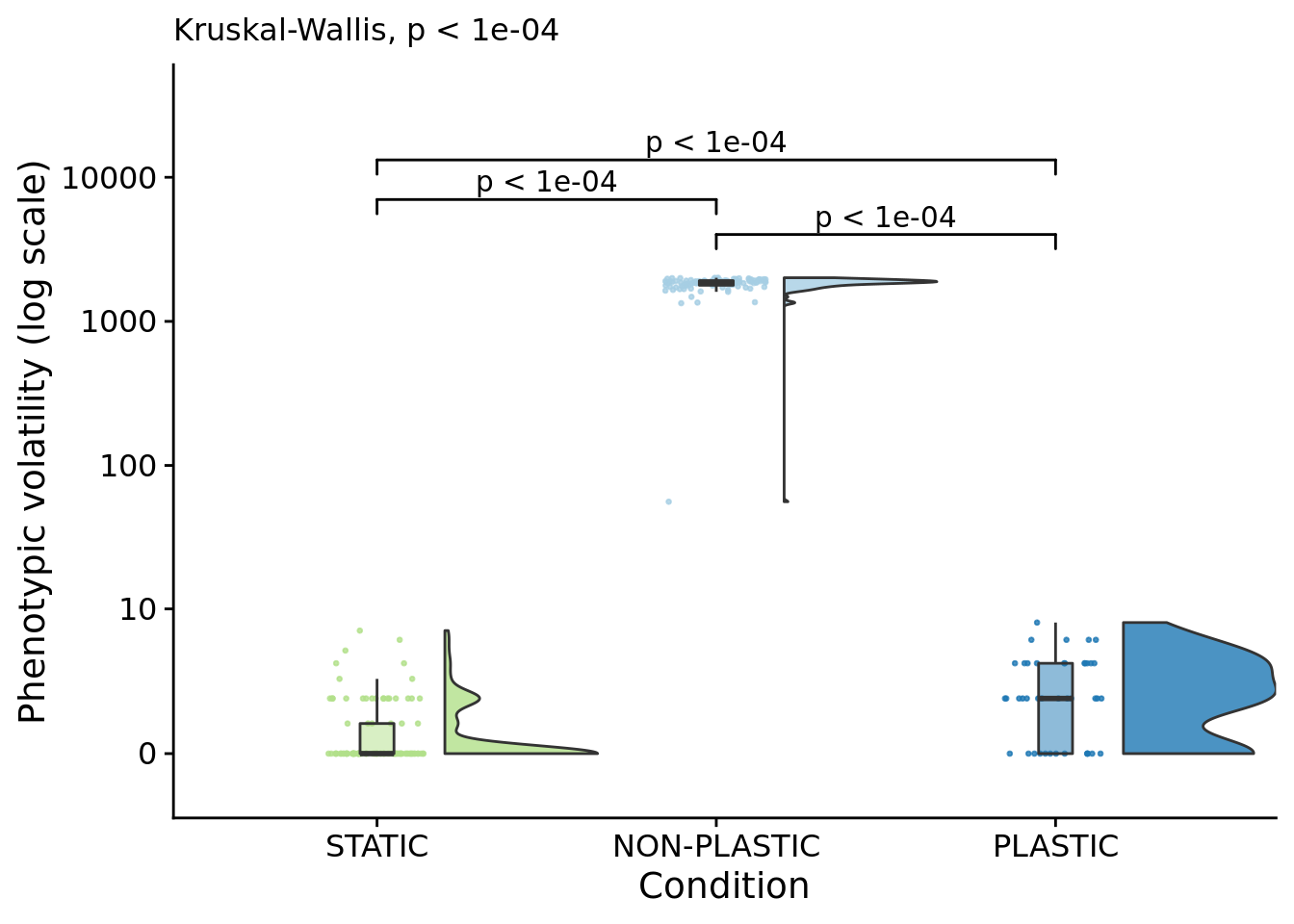
##
## Kruskal-Wallis rank sum test
##
## data: dominant_lineage_trait_volatility by condition
## Kruskal-Wallis chi-squared = 190.78, df = 2, p-value < 2.2e-16pairwise.wilcox.test(
x=summary_data$dominant_lineage_trait_volatility,
g=summary_data$condition,
p.adjust.method="bonferroni",
)##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: summary_data$dominant_lineage_trait_volatility and summary_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC < 2e-16 -
## STATIC < 2e-16 8.7e-07
##
## P value adjustment method: bonferronipaste(
sep="; ",
paste0(
"PLASTIC median: ",
median(filter(summary_data, condition=="PLASTIC")$dominant_lineage_trait_volatility)
),
paste0(
"STATIC median: ",
median(filter(summary_data, condition=="STATIC")$dominant_lineage_trait_volatility)
),
paste0(
"NON-PLASTIC median: ",
median(filter(summary_data, condition=="NON-PLASTIC")$dominant_lineage_trait_volatility)
)
)## [1] "PLASTIC median: 2; STATIC median: 0; NON-PLASTIC median: 1868"## [1] "Wilcox rank sum test statistics:"for (pair in pairwise_comparisons) {
pair_data <- filter(summary_data, condition %in% pair)
pair_data$condition <- as.factor(pair_data$condition)
wt <- wilcox.test(
formula=dominant_lineage_trait_volatility~condition,
data=pair_data,
exact=FALSE,
paired=FALSE
)
print(paste0(pair[1], "<-->", pair[2], ": W=",wt$statistic))
}## [1] "STATIC<-->NON-PLASTIC: W=10000"
## [1] "STATIC<-->PLASTIC: W=3116.5"
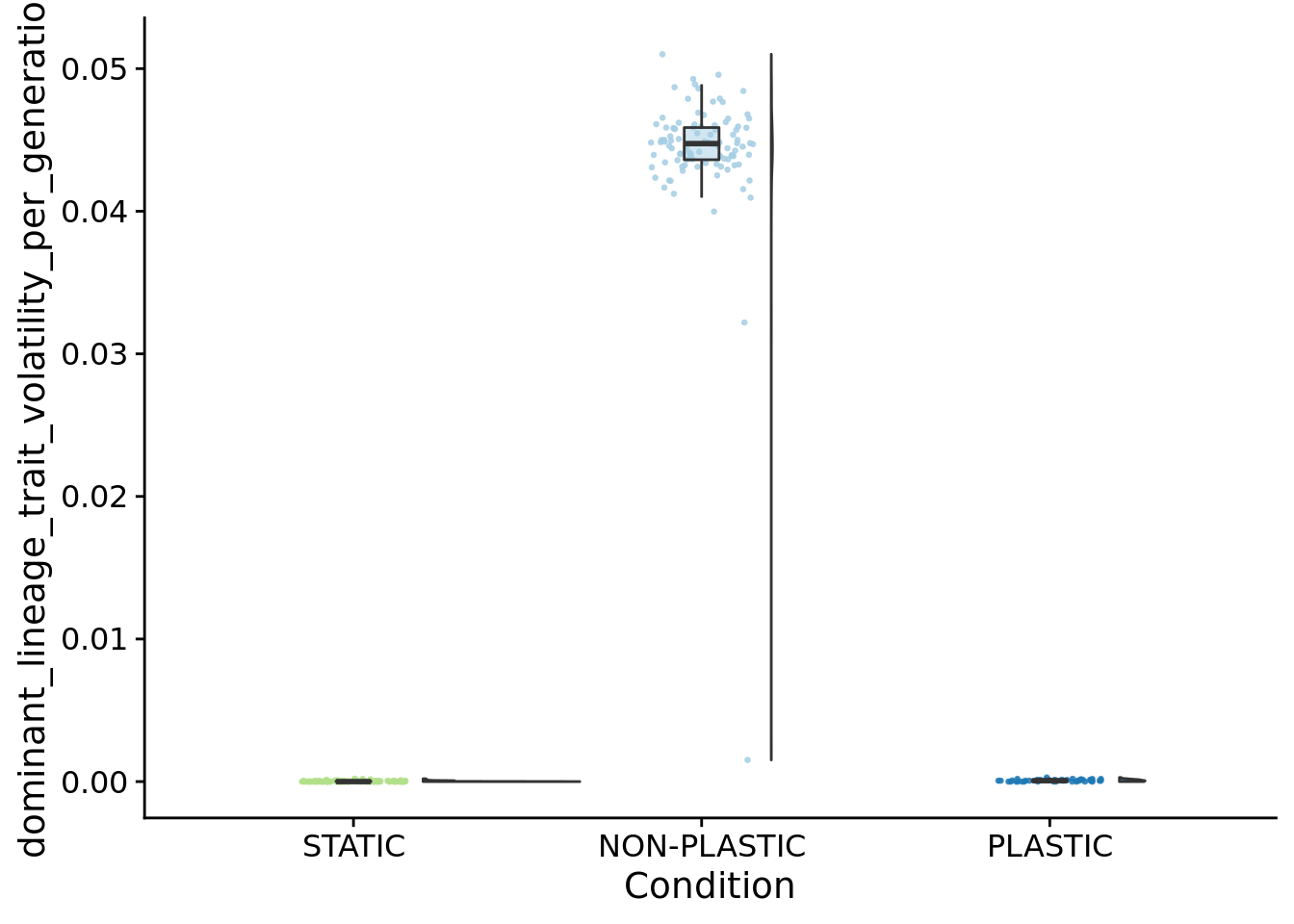
## [1] "PLASTIC<-->NON-PLASTIC: W=4200"6.7.1 Phenotypic volatility normalized by generations elapsed
summary_data$dominant_lineage_trait_volatility_per_generation <- summary_data$dominant_lineage_trait_volatility / summary_data$dominant_generation_born
ggplot(summary_data, aes(x=condition, y=dominant_lineage_trait_volatility_per_generation, fill=condition)) +
geom_flat_violin(
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Condition",
limits=condition_order
) +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
theme(
legend.position="none"
)
kruskal.test(
formula=dominant_lineage_trait_volatility_per_generation~condition,
data=summary_data
)##
## Kruskal-Wallis rank sum test
##
## data: dominant_lineage_trait_volatility_per_generation by condition
## Kruskal-Wallis chi-squared = 189.62, df = 2, p-value < 2.2e-16pairwise.wilcox.test(
x=summary_data$dominant_lineage_trait_volatility_per_generation,
g=summary_data$condition,
p.adjust.method="bonferroni",
)##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: summary_data$dominant_lineage_trait_volatility_per_generation and summary_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC < 2e-16 -
## STATIC < 2e-16 4.2e-06
##
## P value adjustment method: bonferronipaste(
sep="; ",
paste0(
"PLASTIC median: ",
median(filter(summary_data, condition=="PLASTIC")$dominant_lineage_trait_volatility_per_generation)
),
paste0(
"STATIC median: ",
median(filter(summary_data, condition=="STATIC")$dominant_lineage_trait_volatility_per_generation)
),
paste0(
"NON-PLASTIC median: ",
median(filter(summary_data, condition=="NON-PLASTIC")$dominant_lineage_trait_volatility_per_generation)
)
)## [1] "PLASTIC median: 6.33339279717772e-05; STATIC median: 0; NON-PLASTIC median: 0.0447440145638177"## [1] "Wilcox rank sum test statistics:"for (pair in pairwise_comparisons) {
pair_data <- filter(summary_data, condition %in% pair)
pair_data$condition <- as.factor(pair_data$condition)
wt <- wilcox.test(
formula=dominant_lineage_trait_volatility_per_generation~condition,
data=pair_data,
exact=FALSE,
paired=FALSE
)
print(paste0(pair[1], "<-->", pair[2], ": W=",wt$statistic))
}## [1] "STATIC<-->NON-PLASTIC: W=10000"
## [1] "STATIC<-->PLASTIC: W=3061.5"
## [1] "PLASTIC<-->NON-PLASTIC: W=4200"6.8 Phenotypic fidelity
Frequency that an offspring’s genotype is identical to a parent genotype (along the dominant lineage).
summary_data$dominant_lineage_trait_fidelity <- (summary_data$dominant_generation_born - summary_data$dominant_lineage_trait_volatility) / summary_data$dominant_generation_born
# Compute manual labels for geom_signif
stat.test <- summary_data %>%
wilcox_test(dominant_lineage_trait_fidelity ~ condition) %>%
adjust_pvalue(method = "bonferroni") %>%
add_significance() %>%
add_xy_position(x="condition",step.increase=1.5)
# Tweak y.position manually to account for scaled axis (edge case that triggers bad behavior in geom_signif)
stat.test$manual_position <- stat.test$y.position * c(1.0,1.0,1.0005)
stat.test$label <- mapply(p_label,stat.test$p.adj)
summary_data$is_outlier <- mapply(
is_outlier,
summary_data$dominant_lineage_trait_fidelity,
summary_data$condition,
MoreArgs=list(data=summary_data, column="dominant_lineage_trait_fidelity")
)
phenotypic_fidelity_fig <- ggplot(
summary_data,
aes(x=condition, y=dominant_lineage_trait_fidelity, fill=condition)
) +
geom_flat_violin(
data=filter(summary_data,is_outlier==FALSE),
scale="width",
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Condition",
limits=condition_order,
labels=condition_order
) +
scale_y_continuous(
name="Phenotypic fidelity",
limits=c(0.94, 1.013),
breaks=c(0.94, 0.96, 0.98, 1.0) #seq(0.94, 1.0, 0.01)
) +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
# coord_flip() +
labs(
subtitle=paste0(
"Kruskal-Wallis, ",
p_label(signif(kruskal.test(formula=dominant_lineage_trait_fidelity~condition, data=summary_data)$p.value,digits=4))
)
) +
ggsignif::geom_signif(
data=filter(stat.test, p.adj <= alpha),
aes(xmin=group1,xmax=group2,annotations=label,y_position=manual_position),
manual=TRUE,
inherit.aes=FALSE
) +
theme(
legend.position="none"
)## Warning: Ignoring unknown aesthetics: xmin, xmax, annotations, y_positionggsave(
paste0(working_directory, "plots/", "phenotypic-fidelity.pdf"),
width=5,
height=5
)
phenotypic_fidelity_fig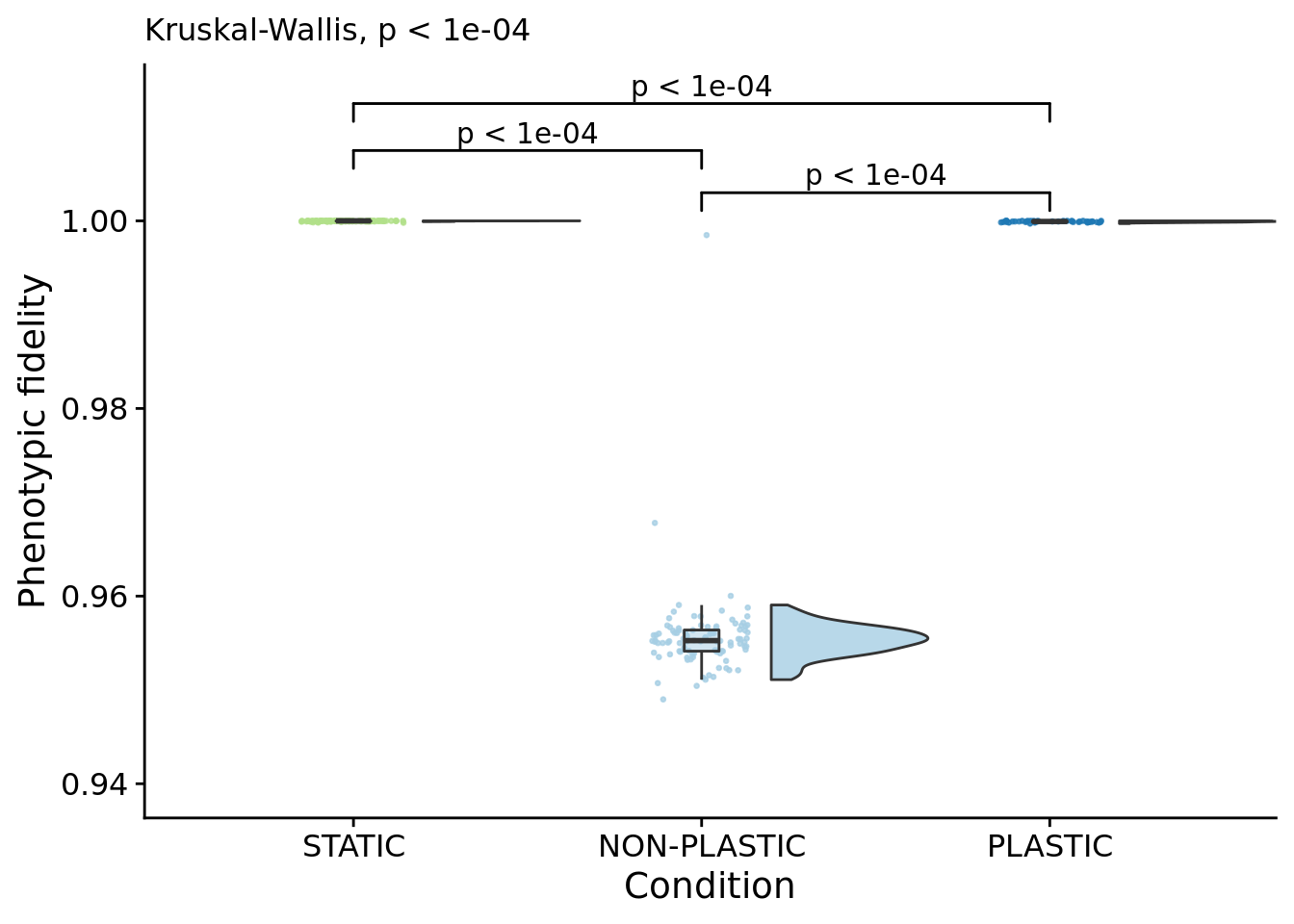
##
## Kruskal-Wallis rank sum test
##
## data: dominant_lineage_trait_fidelity by condition
## Kruskal-Wallis chi-squared = 189.62, df = 2, p-value < 2.2e-16pairwise.wilcox.test(
x=summary_data$dominant_lineage_trait_fidelity,
g=summary_data$condition,
p.adjust.method="bonferroni",
)##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: summary_data$dominant_lineage_trait_fidelity and summary_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC < 2e-16 -
## STATIC < 2e-16 4.2e-06
##
## P value adjustment method: bonferronipaste(
sep="; ",
paste0(
"PLASTIC median: ",
median(filter(summary_data, condition=="PLASTIC")$dominant_lineage_trait_fidelity)
),
paste0(
"STATIC median: ",
median(filter(summary_data, condition=="STATIC")$dominant_lineage_trait_fidelity)
),
paste0(
"NON-PLASTIC median: ",
median(filter(summary_data, condition=="NON-PLASTIC")$dominant_lineage_trait_fidelity)
)
)## [1] "PLASTIC median: 0.999936666072028; STATIC median: 1; NON-PLASTIC median: 0.955255985436182"## [1] "Wilcox rank sum test statistics:"for (pair in pairwise_comparisons) {
pair_data <- filter(summary_data, condition %in% pair)
pair_data$condition <- as.factor(pair_data$condition)
wt <- wilcox.test(
formula=dominant_lineage_trait_fidelity~condition,
data=pair_data,
exact=FALSE,
paired=FALSE
)
print(paste0(pair[1], "<-->", pair[2], ": W=",wt$statistic))
}## [1] "STATIC<-->NON-PLASTIC: W=0"
## [1] "STATIC<-->PLASTIC: W=1138.5"
## [1] "PLASTIC<-->NON-PLASTIC: W=0"6.9 Mutation count
# Compute manual labels for geom_signif
stat.test <- summary_data %>%
wilcox_test(dominant_lineage_total_mut_cnt ~ condition) %>%
adjust_pvalue(method = "bonferroni") %>%
add_significance() %>%
add_xy_position(x="condition",step.increase=1)
# Tweak y.position manually to account for scaled axis (edge case that triggers bad behavior in geom_signif)
stat.test$manual_position <- log10(stat.test$y.position) * c(1.0,1.0,1.03) # c(1.0,1.0,1.01)
stat.test$label <- mapply(p_label,stat.test$p.adj)
summary_data$is_outlier <- mapply(
is_outlier,
summary_data$dominant_lineage_total_mut_cnt,
summary_data$condition,
MoreArgs=list(data=summary_data, column="dominant_lineage_total_mut_cnt")
)
mutation_count_fig <- ggplot(
summary_data,
aes(x=condition, y=dominant_lineage_total_mut_cnt, fill=condition)
) +
geom_flat_violin(
# data=filter(summary_data, !is_outlier),
scale="width",
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Condition",
limits=condition_order,
labels=condition_order
) +
scale_y_continuous(
name="Mutation count (log scale)",
trans=pseudo_log_trans(sigma = 1, base = 10),
breaks=c(0, 10, 100, 1000, 10000),
limits=c(-1, 35000)
) +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
labs(
subtitle=paste0(
"Kruskal-Wallis, ",
p_label(signif(kruskal.test(formula=dominant_lineage_total_mut_cnt~condition, data=summary_data)$p.value,digits=4))
)
) +
ggsignif::geom_signif(
data=filter(stat.test, p.adj <= alpha),
aes(xmin=group1,xmax=group2,annotations=label,y_position=manual_position),
manual=TRUE,
inherit.aes=FALSE
) +
theme(
legend.position="none"
)## Warning: Ignoring unknown aesthetics: xmin, xmax, annotations, y_positionggsave(
paste0(working_directory, "plots/", "mutation-accumulation.pdf"),
width=5,
height=4
)
mutation_count_fig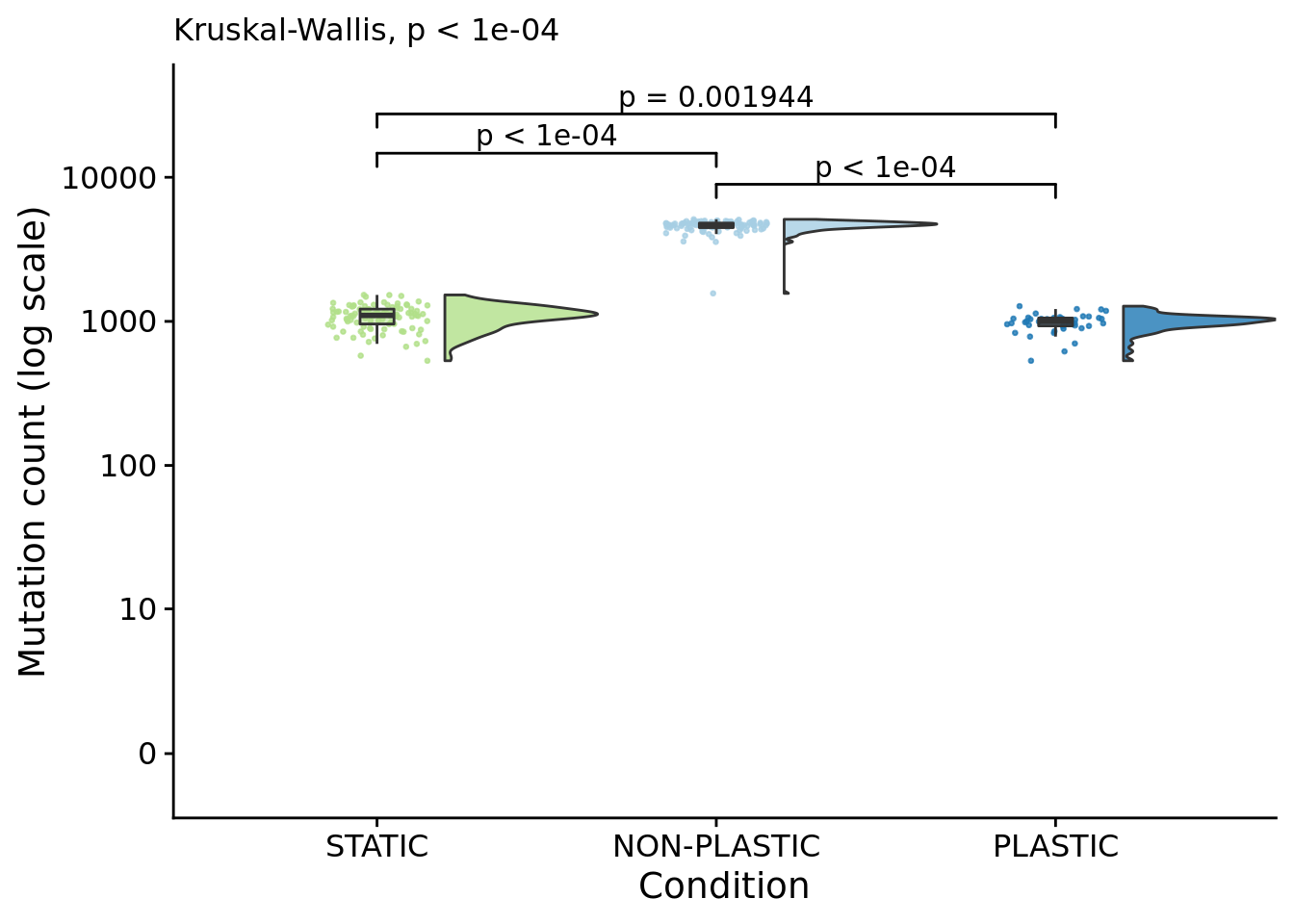
##
## Kruskal-Wallis rank sum test
##
## data: dominant_lineage_total_mut_cnt by condition
## Kruskal-Wallis chi-squared = 179.33, df = 2, p-value < 2.2e-16pairwise.wilcox.test(
x=summary_data$dominant_lineage_total_mut_cnt,
g=summary_data$condition,
p.adjust.method="bonferroni",
)##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: summary_data$dominant_lineage_total_mut_cnt and summary_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC <2e-16 -
## STATIC <2e-16 0.0019
##
## P value adjustment method: bonferronipaste(
sep="; ",
paste0(
"PLASTIC median: ",
median(filter(summary_data, condition=="PLASTIC")$dominant_lineage_total_mut_cnt)
),
paste0(
"STATIC median: ",
median(filter(summary_data, condition=="STATIC")$dominant_lineage_total_mut_cnt)
),
paste0(
"NON-PLASTIC median: ",
median(filter(summary_data, condition=="NON-PLASTIC")$dominant_lineage_total_mut_cnt)
)
)## [1] "PLASTIC median: 998.5; STATIC median: 1100; NON-PLASTIC median: 4657.5"## [1] "Wilcox rank sum test statistics:"for (pair in pairwise_comparisons) {
pair_data <- filter(summary_data, condition %in% pair)
pair_data$condition <- as.factor(pair_data$condition)
wt <- wilcox.test(
formula=dominant_lineage_total_mut_cnt~condition,
data=pair_data,
exact=FALSE,
paired=FALSE
)
print(paste0(pair[1], "<-->", pair[2], ": W=",wt$statistic))
}## [1] "STATIC<-->NON-PLASTIC: W=10000"
## [1] "STATIC<-->PLASTIC: W=1336.5"
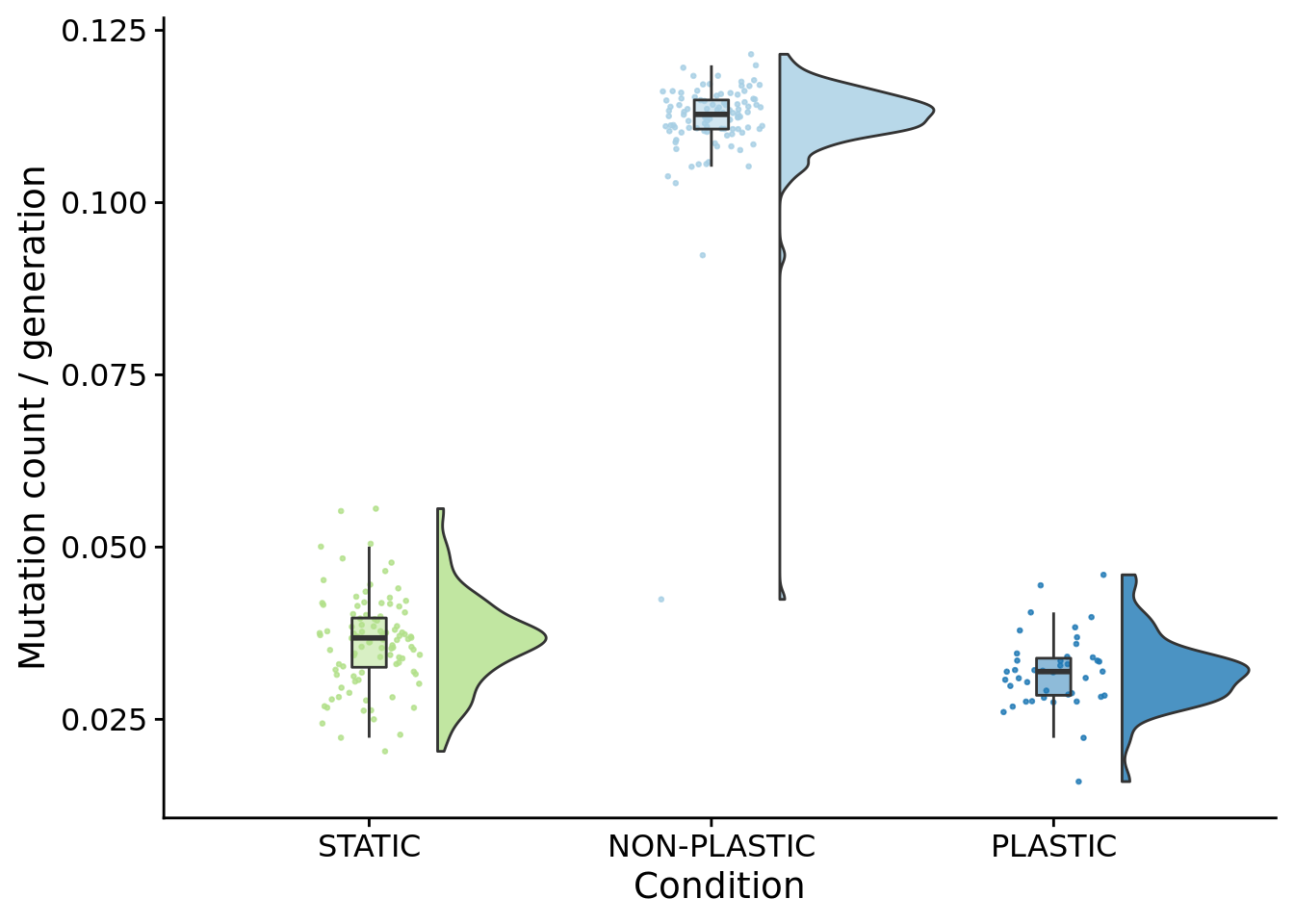
## [1] "PLASTIC<-->NON-PLASTIC: W=4200"6.9.1 Mutation count normalized by generations elapsed
summary_data$mutations_per_generation <- summary_data$dominant_lineage_total_mut_cnt / summary_data$dominant_generation_born
ggplot(summary_data, aes(x=condition, y=mutations_per_generation, fill=condition)) +
geom_flat_violin(
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Condition",
limits=condition_order
) +
ylab("Mutation count / generation") +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
theme(
legend.position="none"
)
##
## Kruskal-Wallis rank sum test
##
## data: mutations_per_generation by condition
## Kruskal-Wallis chi-squared = 180.11, df = 2, p-value < 2.2e-16pairwise.wilcox.test(
x=summary_data$mutations_per_generation,
g=summary_data$condition,
p.adjust.method="bonferroni",
)##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: summary_data$mutations_per_generation and summary_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC <2e-16 -
## STATIC <2e-16 2e-04
##
## P value adjustment method: bonferronipaste(
sep="; ",
paste0(
"PLASTIC median: ",
median(filter(summary_data, condition=="PLASTIC")$mutations_per_generation)
),
paste0(
"STATIC median: ",
median(filter(summary_data, condition=="STATIC")$mutations_per_generation)
),
paste0(
"NON-PLASTIC median: ",
median(filter(summary_data, condition=="NON-PLASTIC")$mutations_per_generation)
)
)## [1] "PLASTIC median: 0.0319267181456982; STATIC median: 0.0368157192941933; NON-PLASTIC median: 0.112804526786948"## [1] "Wilcox rank sum test statistics:"for (pair in pairwise_comparisons) {
pair_data <- filter(summary_data, condition %in% pair)
pair_data$condition <- as.factor(pair_data$condition)
wt <- wilcox.test(
formula=mutations_per_generation~condition,
data=pair_data,
exact=FALSE,
paired=FALSE
)
print(paste0(pair[1], "<-->", pair[2], ": W=",wt$statistic))
}## [1] "STATIC<-->NON-PLASTIC: W=9987"
## [1] "STATIC<-->PLASTIC: W=1206"
## [1] "PLASTIC<-->NON-PLASTIC: W=4198"6.10 Genotypic fidelity
The frequency that an offspring’s genotype is the same as a parent’s genotype.
summary_data$dominant_lineage_genotypic_fidelity <- (summary_data$dominant_generation_born - summary_data$dominant_lineage_num_mut_steps) / summary_data$dominant_generation_born
# Compute manual labels for geom_signif
stat.test <- summary_data %>%
wilcox_test(dominant_lineage_genotypic_fidelity ~ condition) %>%
adjust_pvalue(method = "bonferroni") %>%
add_significance() %>%
add_xy_position(x="condition",step.increase=0.2)
# Tweak y.position manually to account for scaled axis (edge case that triggers bad behavior in geom_signif)
stat.test$manual_position <- stat.test$y.position * c(1.0,1.0,1.0)
stat.test$label <- mapply(p_label,stat.test$p.adj)
genotypic_fidelity_fig <- ggplot(
summary_data,
aes(x=condition, y=dominant_lineage_genotypic_fidelity, fill=condition)
) +
geom_flat_violin(
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Condition",
limits=condition_order,
labels=condition_order
) +
scale_y_continuous(
name="Genotypic fidelity",
limits=c(0.85, 1.01),
breaks=c(0.85, 0.90, 0.95, 1.0) #seq(0.85, 1.0, 0.02)
) +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
# coord_flip() +
labs(
subtitle=paste0(
"Kruskal-Wallis, ",
p_label(signif(kruskal.test(formula=dominant_lineage_genotypic_fidelity~condition, data=summary_data)$p.value,digits=4))
)
) +
ggsignif::geom_signif(
data=filter(stat.test, p.adj <= alpha),
aes(xmin=group1,xmax=group2,annotations=label,y_position=manual_position),
manual=TRUE,
inherit.aes=FALSE
) +
theme(
legend.position="none"
)## Warning: Ignoring unknown aesthetics: xmin, xmax, annotations, y_positionggsave(
paste0(working_directory, "plots/", "genotypic-fidelity.pdf"),
width=5,
height=4
)
genotypic_fidelity_fig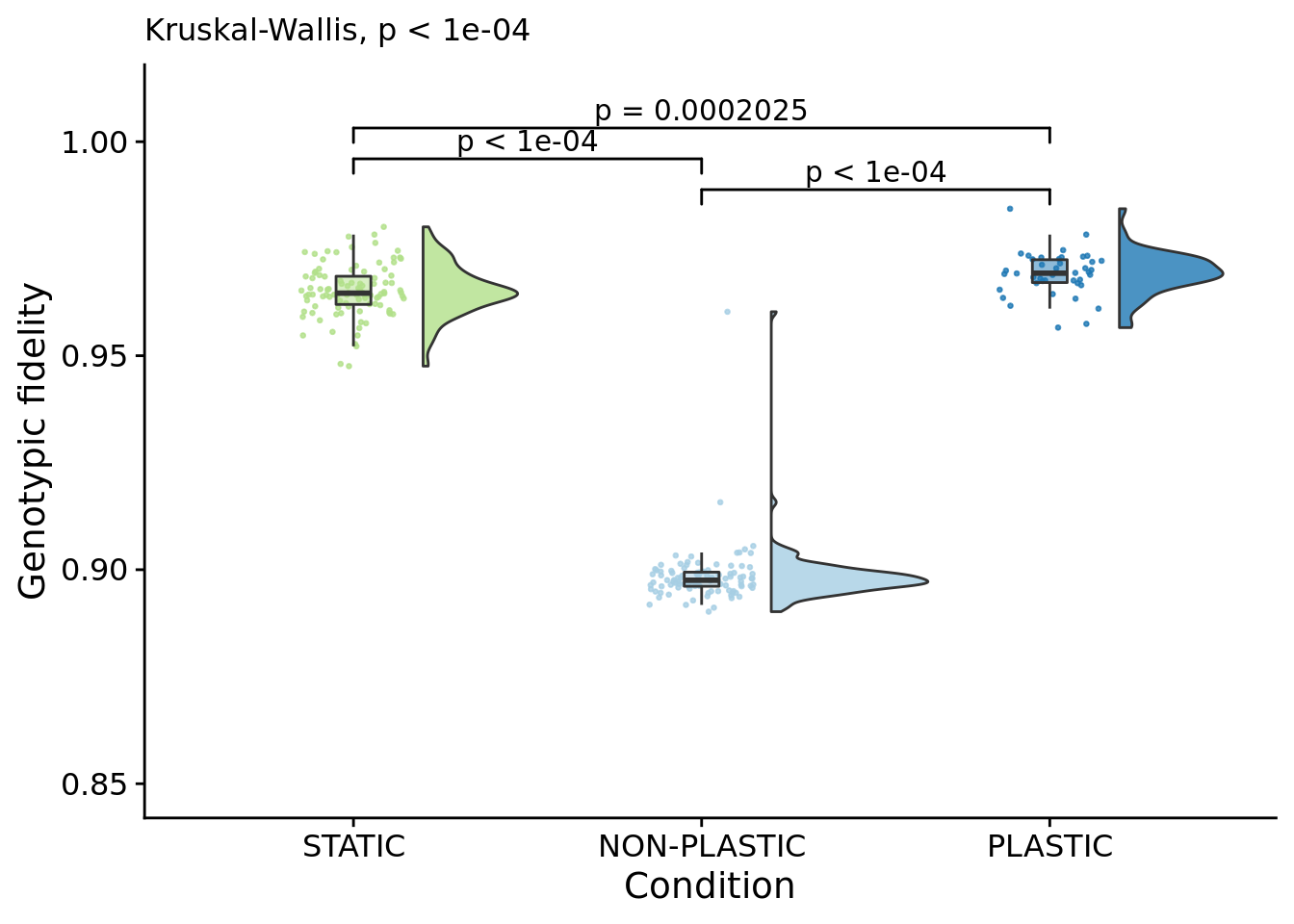
##
## Kruskal-Wallis rank sum test
##
## data: dominant_lineage_genotypic_fidelity by condition
## Kruskal-Wallis chi-squared = 179.86, df = 2, p-value < 2.2e-16pairwise.wilcox.test(
x=summary_data$dominant_lineage_genotypic_fidelity,
g=summary_data$condition,
p.adjust.method="bonferroni",
)##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: summary_data$dominant_lineage_genotypic_fidelity and summary_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC <2e-16 -
## STATIC <2e-16 2e-04
##
## P value adjustment method: bonferronipaste(
sep="; ",
paste0(
"PLASTIC median: ",
median(filter(summary_data, condition=="PLASTIC")$dominant_lineage_genotypic_fidelity)
),
paste0(
"STATIC median: ",
median(filter(summary_data, condition=="STATIC")$dominant_lineage_genotypic_fidelity)
),
paste0(
"NON-PLASTIC median: ",
median(filter(summary_data, condition=="NON-PLASTIC")$dominant_lineage_genotypic_fidelity)
)
)## [1] "PLASTIC median: 0.969286906891951; STATIC median: 0.964620594632577; NON-PLASTIC median: 0.89754902563783"## [1] "Wilcox rank sum test statistics:"for (pair in pairwise_comparisons) {
pair_data <- filter(summary_data, condition %in% pair)
pair_data$condition <- as.factor(pair_data$condition)
wt <- wilcox.test(
formula=dominant_lineage_genotypic_fidelity~condition,
data=pair_data,
exact=FALSE,
paired=FALSE
)
print(paste0(pair[1], "<-->", pair[2], ": W=",wt$statistic))
}## [1] "STATIC<-->NON-PLASTIC: W=18"
## [1] "STATIC<-->PLASTIC: W=2992"
## [1] "PLASTIC<-->NON-PLASTIC: W=2"6.11 Characterizing variation along dominant lineages
6.11.1 Mutational instability
summary_data$frac_phenotype_changing_mut_steps <- summary_data$dominant_lineage_num_mut_steps_that_change_aggregate_phenotype / summary_data$dominant_lineage_num_mut_steps
summary_data$frac_phenotype_stable_mut_steps <- 1 - summary_data$frac_phenotype_changing_mut_steps
# Compute manual labels for geom_signif
stat.test <- summary_data %>%
wilcox_test(frac_phenotype_changing_mut_steps ~ condition) %>%
adjust_pvalue(method = "bonferroni") %>%
add_significance() %>%
add_xy_position(x="condition",step.increase=0.2)
# Tweak y.position manually to account for scaled axis (edge case that triggers bad behavior in geom_signif)
stat.test$manual_position <- stat.test$y.position #* c(1.0,1.0,1.0)
stat.test$label <- mapply(p_label,stat.test$p.adj)
ggplot(summary_data, aes(x=condition, y=frac_phenotype_changing_mut_steps, fill=condition)) +
geom_flat_violin(
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Condition",
limits=condition_order
) +
ylab("Mutational instability") +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
# coord_flip() +
labs(
subtitle=paste0(
"Kruskal-Wallis, ",
p_label(signif(kruskal.test(formula=frac_phenotype_changing_mut_steps~condition, data=summary_data)$p.value,digits=4))
)
) +
ggsignif::geom_signif(
data=filter(stat.test, p.adj <= alpha),
aes(xmin=group1,xmax=group2,annotations=label,y_position=manual_position),
manual=TRUE,
inherit.aes=FALSE
) +
theme(
legend.position="none"
)## Warning: Ignoring unknown aesthetics: xmin, xmax, annotations, y_position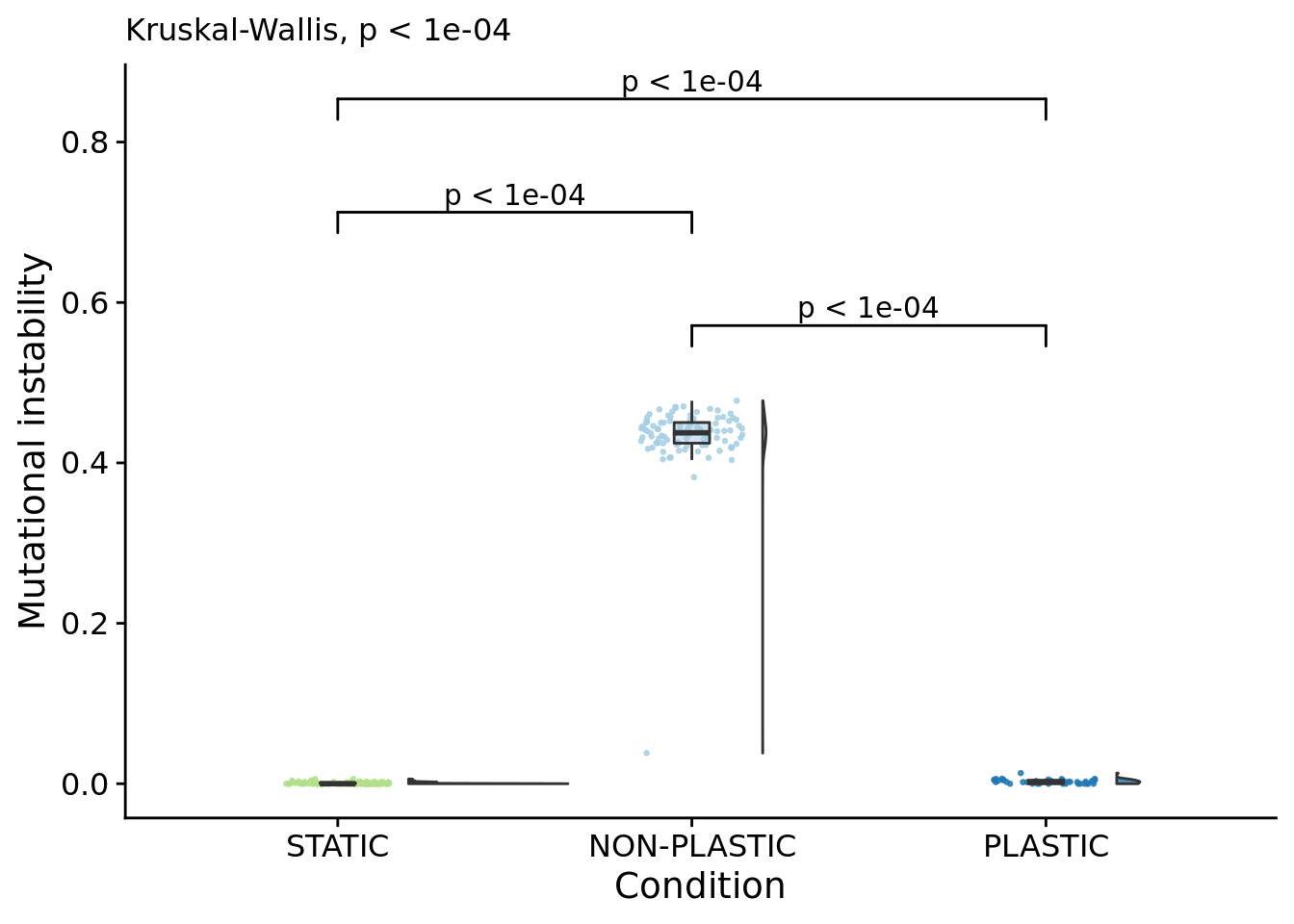
## Saving 7 x 5 in image##
## Kruskal-Wallis rank sum test
##
## data: frac_phenotype_changing_mut_steps by condition
## Kruskal-Wallis chi-squared = 191.23, df = 2, p-value < 2.2e-16pairwise.wilcox.test(
x=summary_data$frac_phenotype_changing_mut_steps,
g=summary_data$condition,
p.adjust.method="bonferroni",
)##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: summary_data$frac_phenotype_changing_mut_steps and summary_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC < 2e-16 -
## STATIC < 2e-16 2.3e-07
##
## P value adjustment method: bonferronipaste(
sep="; ",
paste0(
"PLASTIC median: ",
median(filter(summary_data, condition=="PLASTIC")$frac_phenotype_changing_mut_steps)
),
paste0(
"STATIC median: ",
median(filter(summary_data, condition=="STATIC")$frac_phenotype_changing_mut_steps)
),
paste0(
"NON-PLASTIC median: ",
median(filter(summary_data, condition=="NON-PLASTIC")$frac_phenotype_changing_mut_steps)
)
)## [1] "PLASTIC median: 0.00224941742616098; STATIC median: 0; NON-PLASTIC median: 0.437583018324547"## [1] "Wilcox rank sum test statistics:"for (pair in pairwise_comparisons) {
pair_data <- filter(summary_data, condition %in% pair)
pair_data$condition <- as.factor(pair_data$condition)
wt <- wilcox.test(
formula=frac_phenotype_changing_mut_steps~condition,
data=pair_data,
exact=FALSE,
paired=FALSE
)
print(paste0(pair[1], "<-->", pair[2], ": W=",wt$statistic))
}## [1] "STATIC<-->NON-PLASTIC: W=10000"
## [1] "STATIC<-->PLASTIC: W=3172"
## [1] "PLASTIC<-->NON-PLASTIC: W=4200"6.11.2 Mutational stability (realized mutational robustness)
# Compute manual labels for geom_signif
stat.test <- summary_data %>%
wilcox_test(frac_phenotype_stable_mut_steps ~ condition) %>%
adjust_pvalue(method = "bonferroni") %>%
add_significance() %>%
add_xy_position(x="condition",step.increase=0.75)
# Tweak y.position manually to account for scaled axis (edge case that triggers bad behavior in geom_signif)
stat.test$manual_position <- stat.test$y.position #* c(1.0,1.0,1.0)
stat.test$label <- mapply(p_label,stat.test$p.adj)
summary_data$is_outlier <- mapply(
is_outlier,
summary_data$dominant_lineage_trait_volatility,
summary_data$condition,
MoreArgs=list(data=summary_data, column="dominant_lineage_trait_volatility")
)
mutational_stability_fig <- ggplot(
summary_data,
aes(x=condition, y=frac_phenotype_stable_mut_steps, fill=condition)
) +
geom_flat_violin(
# data=filter(summary_data,is_outlier==FALSE),
scale="width",
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Condition",
limits=condition_order
) +
scale_y_continuous(
name="Realized mutational robustness",
limits=c(0.5, 1.15),
breaks=c(0.5, 0.75, 1.0)
) +
scale_fill_brewer(
palette=cb_palette
) +
scale_color_brewer(
palette=cb_palette
) +
labs(
subtitle=paste0(
"Kruskal-Wallis, ",
p_label(signif(kruskal.test(formula=frac_phenotype_stable_mut_steps~condition, data=summary_data)$p.value,digits=4))
)
) +
ggsignif::geom_signif(
data=filter(stat.test, p.adj <= alpha),
aes(xmin=group1,xmax=group2,annotations=label,y_position=manual_position),
manual=TRUE,
inherit.aes=FALSE
) +
theme(
legend.position="none"
)## Warning: Ignoring unknown aesthetics: xmin, xmax, annotations, y_position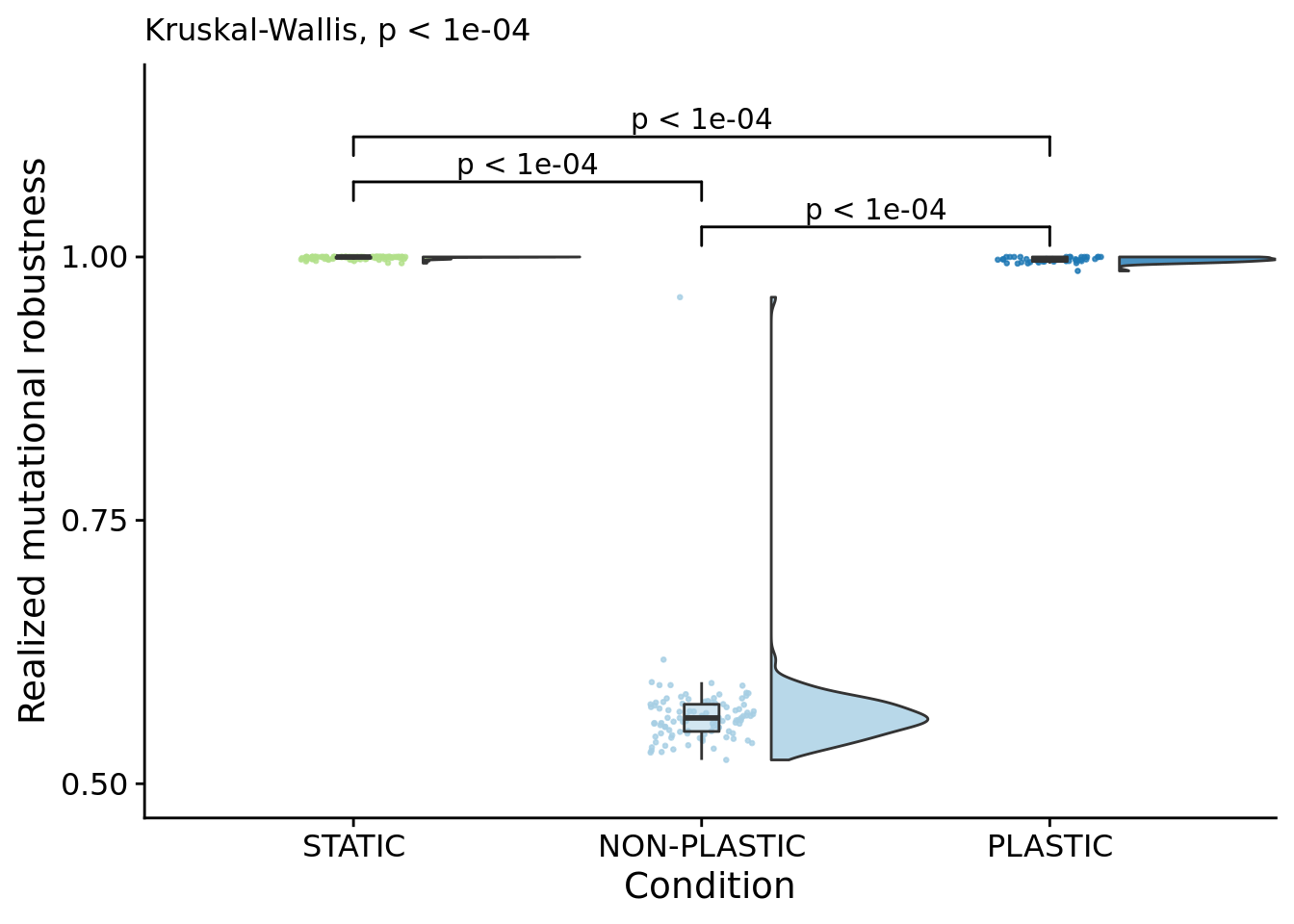
##
## Kruskal-Wallis rank sum test
##
## data: frac_phenotype_stable_mut_steps by condition
## Kruskal-Wallis chi-squared = 191.23, df = 2, p-value < 2.2e-16pairwise.wilcox.test(
x=summary_data$frac_phenotype_stable_mut_steps,
g=summary_data$condition,
p.adjust.method="bonferroni",
)##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: summary_data$frac_phenotype_stable_mut_steps and summary_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC < 2e-16 -
## STATIC < 2e-16 2.3e-07
##
## P value adjustment method: bonferronipaste(
sep="; ",
paste0(
"PLASTIC median: ",
median(filter(summary_data, condition=="PLASTIC")$frac_phenotype_stable_mut_steps)
),
paste0(
"STATIC median: ",
median(filter(summary_data, condition=="STATIC")$frac_phenotype_stable_mut_steps)
),
paste0(
"NON-PLASTIC median: ",
median(filter(summary_data, condition=="NON-PLASTIC")$frac_phenotype_stable_mut_steps)
)
)## [1] "PLASTIC median: 0.997750582573839; STATIC median: 1; NON-PLASTIC median: 0.562416981675453"## [1] "Wilcox rank sum test statistics:"for (pair in pairwise_comparisons) {
pair_data <- filter(summary_data, condition %in% pair)
pair_data$condition <- as.factor(pair_data$condition)
wt <- wilcox.test(
formula=frac_phenotype_stable_mut_steps~condition,
data=pair_data,
exact=FALSE,
paired=FALSE
)
print(paste0(pair[1], "<-->", pair[2], ": W=",wt$statistic))
}## [1] "STATIC<-->NON-PLASTIC: W=0"
## [1] "STATIC<-->PLASTIC: W=1028"
## [1] "PLASTIC<-->NON-PLASTIC: W=0"6.11.3 For PLASTIC populations, what fraction of phenotype-altering mutations occurred in the unexpressed phenotype?
summary_data$frac_unexpressed_mut_steps <- summary_data$dominant_lineage_num_mut_steps_that_change_unexpressed_phenotype / summary_data$dominant_lineage_num_mut_steps_that_change_aggregate_phenotype
summary_data$frac_expressed_mut_steps <- summary_data$dominant_lineage_num_mut_steps_that_change_expressed_phenotype / summary_data$dominant_lineage_num_mut_steps_that_change_aggregate_phenotype
ggplot(filter(summary_data, condition=="PLASTIC" & dominant_lineage_num_mut_steps_that_change_aggregate_phenotype > 0), aes(x=frac_unexpressed_mut_steps)) +
geom_histogram(binwidth=0.1) +
scale_x_continuous(
limits=c(0, 1.1),
breaks=seq(0, 1.0, 0.1)
) +
theme(
legend.position="none"
)## Warning: Removed 2 rows containing missing values (geom_bar).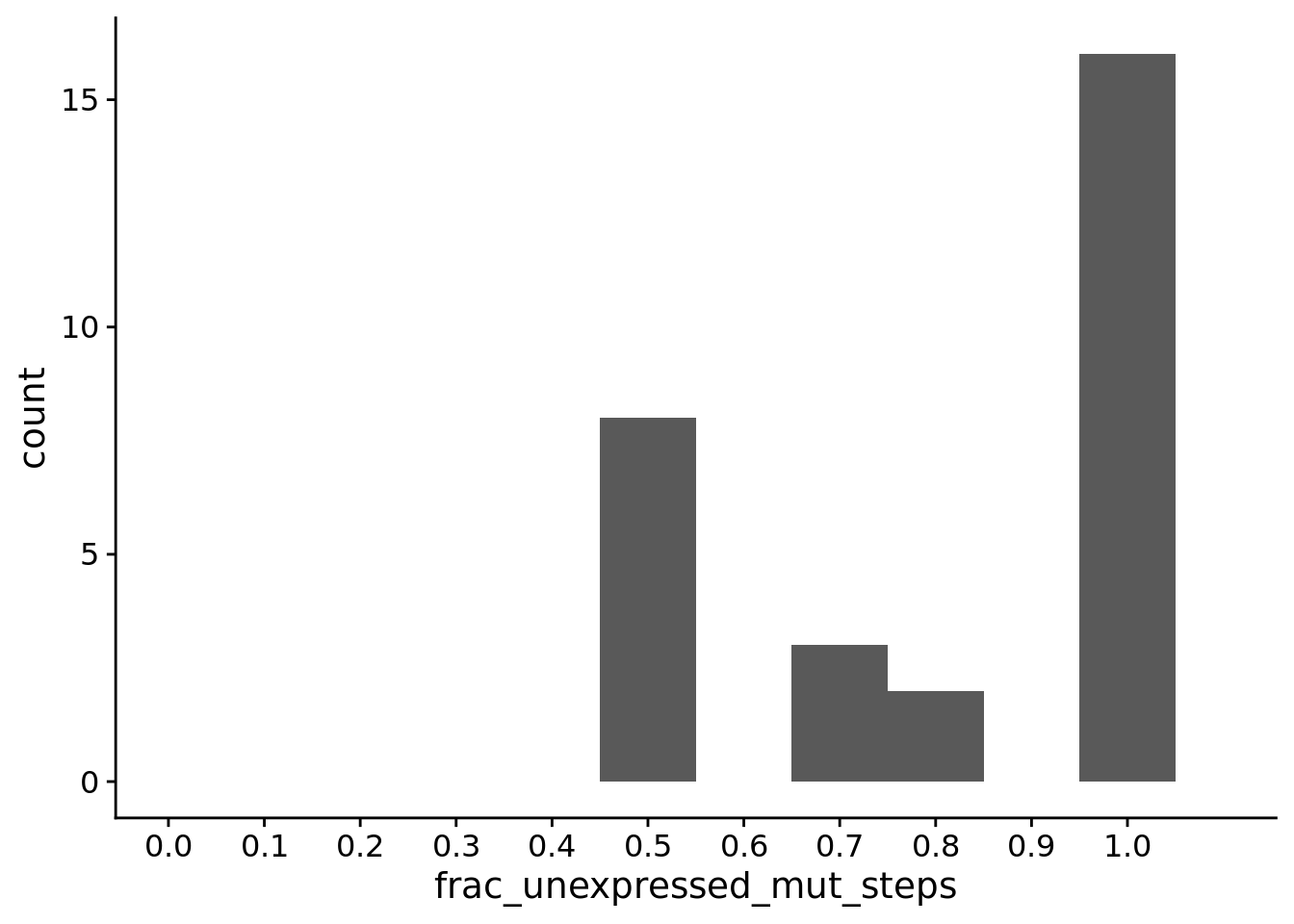
## [1] "PLASTIC - Mean with bootstrapped 95% CI"bo <- boot(filter(summary_data, condition=="PLASTIC" & dominant_lineage_num_mut_steps_that_change_aggregate_phenotype > 0)$frac_unexpressed_mut_steps, statistic=samplemean, R=10000)
print(bo)##
## ORDINARY NONPARAMETRIC BOOTSTRAP
##
##
## Call:
## boot(data = filter(summary_data, condition == "PLASTIC" & dominant_lineage_num_mut_steps_that_change_aggregate_phenotype >
## 0)$frac_unexpressed_mut_steps, statistic = samplemean, R = 10000)
##
##
## Bootstrap Statistics :
## original bias std. error
## t1* 0.8247126 0.0002471264 0.03986957## BOOTSTRAP CONFIDENCE INTERVAL CALCULATIONS
## Based on 10000 bootstrap replicates
##
## CALL :
## boot.ci(boot.out = bo, conf = 0.95, type = "perc")
##
## Intervals :
## Level Percentile
## 95% ( 0.7443, 0.8994 )
## Calculations and Intervals on Original Scaleplastic_summary_data <- filter(summary_data, condition=="PLASTIC")
aggregate_frac_mut_steps_that_change_unexpressed_phenotype <- sum(plastic_summary_data$dominant_lineage_num_mut_steps_that_change_unexpressed_phenotype) / sum(plastic_summary_data$dominant_lineage_num_mut_steps_that_change_aggregate_phenotype)
sum(plastic_summary_data$dominant_lineage_num_mut_steps_that_change_unexpressed_phenotype)## [1] 83## [1] 102## [1] 0.813725583 / 102 (0.8137255)
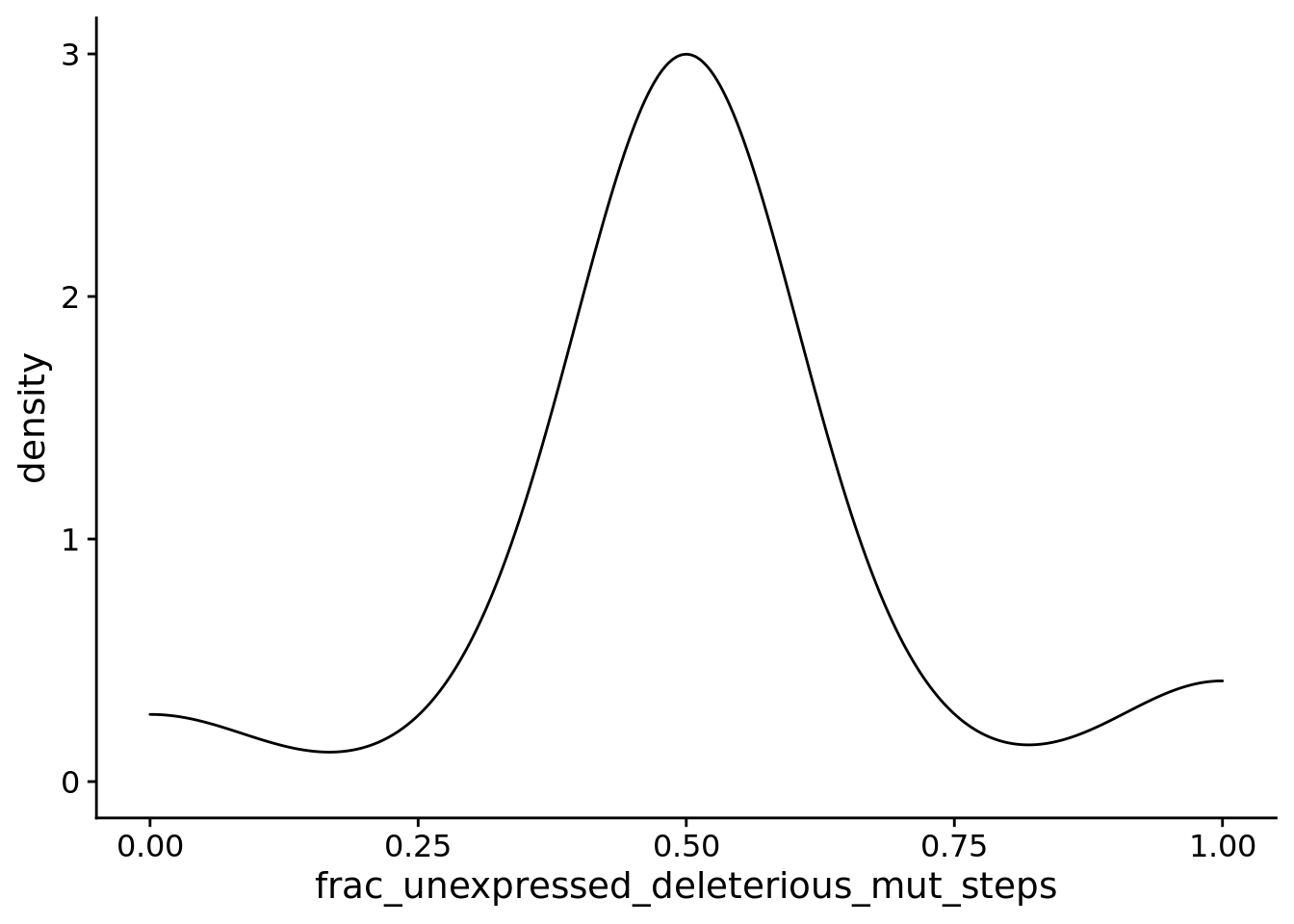
6.11.4 For PLASTIC populations, what fraction of mutations that affect the unexpressed phenotype are deleterious versus beneficial?
aggregate_frac_unexpressed_deleterious_mut_steps <- sum(plastic_summary_data$dominant_lineage_num_mut_steps_that_change_unexpressed_phenotype_deleterious) / sum(plastic_summary_data$dominant_lineage_num_mut_steps_that_change_unexpressed_phenotype)
aggregate_frac_unexpressed_beneficial_mut_steps <- sum(plastic_summary_data$dominant_lineage_num_mut_steps_that_change_unexpressed_phenotype_beneficial) / sum(plastic_summary_data$dominant_lineage_num_mut_steps_that_change_unexpressed_phenotype)6.11.4.1 Deleterious mutations
summary_data$frac_unexpressed_deleterious_mut_steps <- summary_data$dominant_lineage_num_mut_steps_that_change_unexpressed_phenotype_deleterious / summary_data$dominant_lineage_num_mut_steps_that_change_unexpressed_phenotype
ggplot(
filter(summary_data, condition=="PLASTIC" & dominant_lineage_num_mut_steps_that_change_unexpressed_phenotype > 0),
aes(x=frac_unexpressed_deleterious_mut_steps)
) +
geom_density() +
theme(
legend.position="none"
)
bo <- boot(filter(summary_data, condition=="PLASTIC" & dominant_lineage_num_mut_steps_that_change_aggregate_phenotype > 0)$frac_unexpressed_deleterious_mut_steps, statistic=samplemean, R=10000)
print(bo)##
## ORDINARY NONPARAMETRIC BOOTSTRAP
##
##
## Call:
## boot(data = filter(summary_data, condition == "PLASTIC" & dominant_lineage_num_mut_steps_that_change_aggregate_phenotype >
## 0)$frac_unexpressed_deleterious_mut_steps, statistic = samplemean,
## R = 10000)
##
##
## Bootstrap Statistics :
## original bias std. error
## t1* 0.5172414 -0.0004291954 0.0395998## BOOTSTRAP CONFIDENCE INTERVAL CALCULATIONS
## Based on 10000 bootstrap replicates
##
## CALL :
## boot.ci(boot.out = bo, conf = 0.95, type = "perc")
##
## Intervals :
## Level Percentile
## 95% ( 0.4391, 0.5954 )
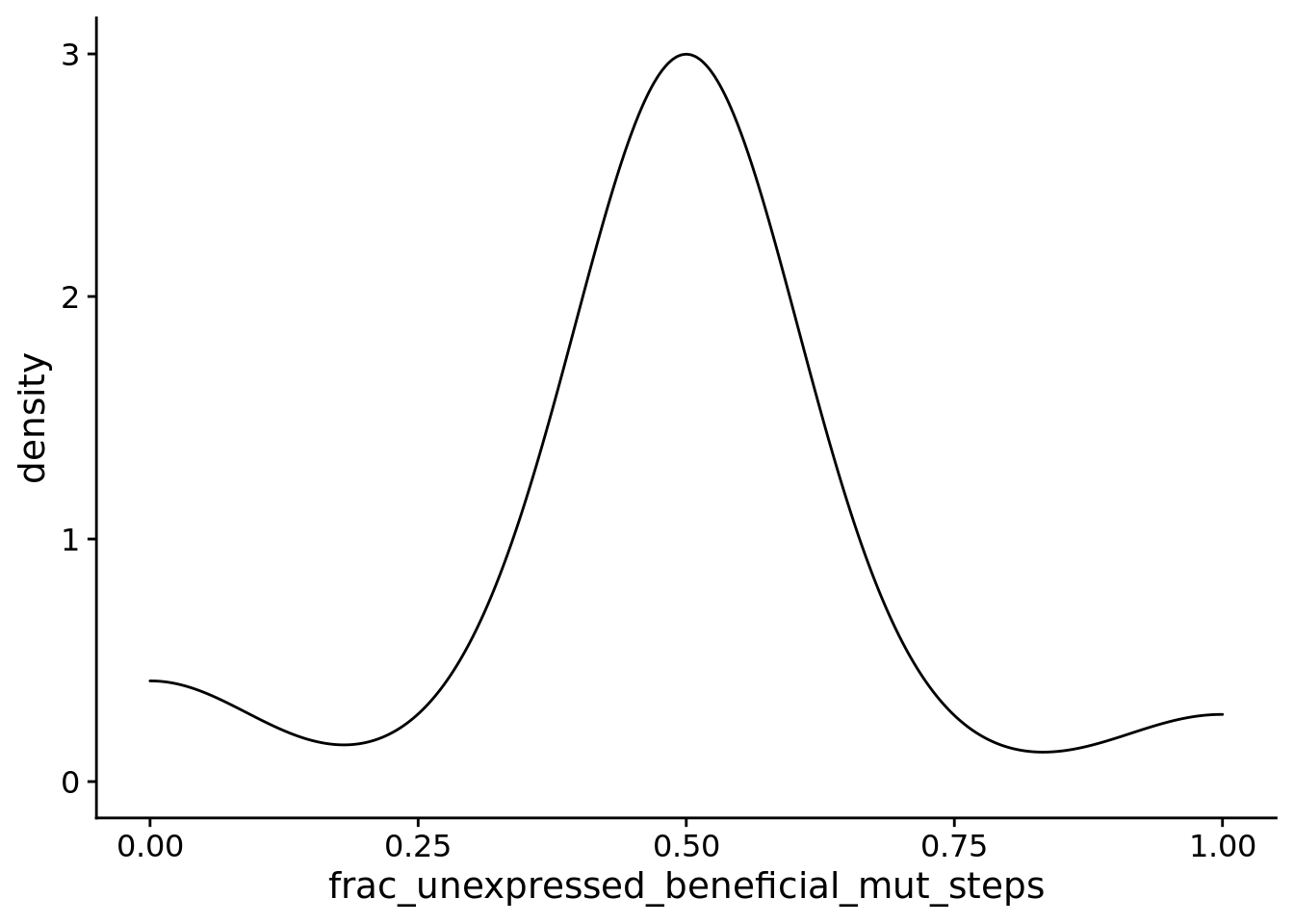
## Calculations and Intervals on Original Scale6.11.4.2 Beneficial mutations
summary_data$frac_unexpressed_beneficial_mut_steps <- summary_data$dominant_lineage_num_mut_steps_that_change_unexpressed_phenotype_beneficial / summary_data$dominant_lineage_num_mut_steps_that_change_unexpressed_phenotype
ggplot(
filter(summary_data, condition=="PLASTIC" & dominant_lineage_num_mut_steps_that_change_unexpressed_phenotype > 0),
aes(x=frac_unexpressed_beneficial_mut_steps)
) +
geom_density() +
theme(
legend.position="none"
)
bo <- boot(filter(summary_data, condition=="PLASTIC" & dominant_lineage_num_mut_steps_that_change_aggregate_phenotype > 0)$frac_unexpressed_beneficial_mut_steps, statistic=samplemean, R=10000)
print(bo)##
## ORDINARY NONPARAMETRIC BOOTSTRAP
##
##
## Call:
## boot(data = filter(summary_data, condition == "PLASTIC" & dominant_lineage_num_mut_steps_that_change_aggregate_phenotype >
## 0)$frac_unexpressed_beneficial_mut_steps, statistic = samplemean,
## R = 10000)
##
##
## Bootstrap Statistics :
## original bias std. error
## t1* 0.4827586 -9.436782e-05 0.03874561## BOOTSTRAP CONFIDENCE INTERVAL CALCULATIONS
## Based on 10000 bootstrap replicates
##
## CALL :
## boot.ci(boot.out = bo, conf = 0.95, type = "perc")
##
## Intervals :
## Level Percentile
## 95% ( 0.4046, 0.5586 )
## Calculations and Intervals on Original Scale6.12 Mutational robustness
Mutational robustness measures the fraction of one-step mutations on a focal genotype that result in a phenotypic change. Here, we calculate the mutational robustness of the representative genotype from each replicate (the most abundant genotype in the final population).
This data is located in a separate .tar.gz file on OSF, so we need to load it and wrangle the data.
# Load the data
df_mut = read.csv(paste0(working_directory, 'mutational_robustness/data/aggregated_mutant_data.csv'))
# Extract the treatment for each line
df_mut$treatment = 'STATIC'
df_mut[df_mut$environment == 'chg-u100',]$treatment = 'PLASTIC'
df_mut[df_mut$environment == 'chg-u100' & df_mut$sensors == F,]$treatment = 'NON-PLASTIC'
df_mut$treatment_factor = factor(df_mut$treatment, levels = c('STATIC', 'NON-PLASTIC', 'PLASTIC'))
# For compatibility with is_outlier above
df_mut$condition = df_mut$treatment_factor
# Calculate robustness (originally calculated as volatility)
df_mut$mutational_robustness = 1 - df_mut$one_step_diff_pheno_fracNow we can plot mutational robustness:
# Compute manual labels for geom_signif
stat.test <- df_mut %>%
wilcox_test(mutational_robustness ~ treatment_factor) %>%
adjust_pvalue(method = "bonferroni") %>%
add_significance() %>%
add_xy_position(x="treatment_factor")
# Tweak y.position manually to account for scaled axis (edge case that triggers bad behavior in geom_signif)
stat.test$manual_position <- stat.test$y.position * c(1.05,1.1,1.15)
stat.test$label <- mapply(p_label,stat.test$p.adj)
df_mut$is_outlier <- mapply(
is_outlier,
df_mut$mutational_robustness,
df_mut$treatment_factor,
MoreArgs=list(data=df_mut, column="mutational_robustness")
)
# Remap colors so that colors map
color_map = c(
'STATIC' = brewer.pal(3, cb_palette)[3],
'PLASTIC' = brewer.pal(3, cb_palette)[2],
'NON-PLASTIC' = brewer.pal(3, cb_palette)[1]
)
# Plot!
mut_robustness_fig <- ggplot(df_mut, aes(x=treatment_factor, y=mutational_robustness, fill=treatment_factor)) +
geom_flat_violin( data=filter(df_mut,is_outlier==FALSE),
scale="width", position = position_nudge(x = .2, y = 0), alpha = .8) +
geom_point(mapping=aes(color=treatment_factor), position = position_jitter(width = .15), size = .5, alpha = 0.8) +
geom_boxplot(width = .1, outlier.shape = NA, alpha = 0.5) +
scale_x_discrete( name="Condition") +
scale_y_continuous( name='Mutational robustness', limits=c(0, 1)) +
scale_fill_manual( values = color_map ) +
scale_color_manual( values = color_map ) +
labs( subtitle=paste0( "Kruskal-Wallis, ", p_label(signif(kruskal.test(formula=mutational_robustness~treatment_factor, data=df_mut)$p.value,digits=5)) ) ) +
ggsignif::geom_signif( data=filter(stat.test, p.adj <= alpha), aes(xmin=group1,xmax=group2,annotations=label,y_position=manual_position), manual=TRUE, inherit.aes=FALSE ) +
theme( legend.position="none" )## Warning: Ignoring unknown aesthetics: xmin, xmax, annotations, y_position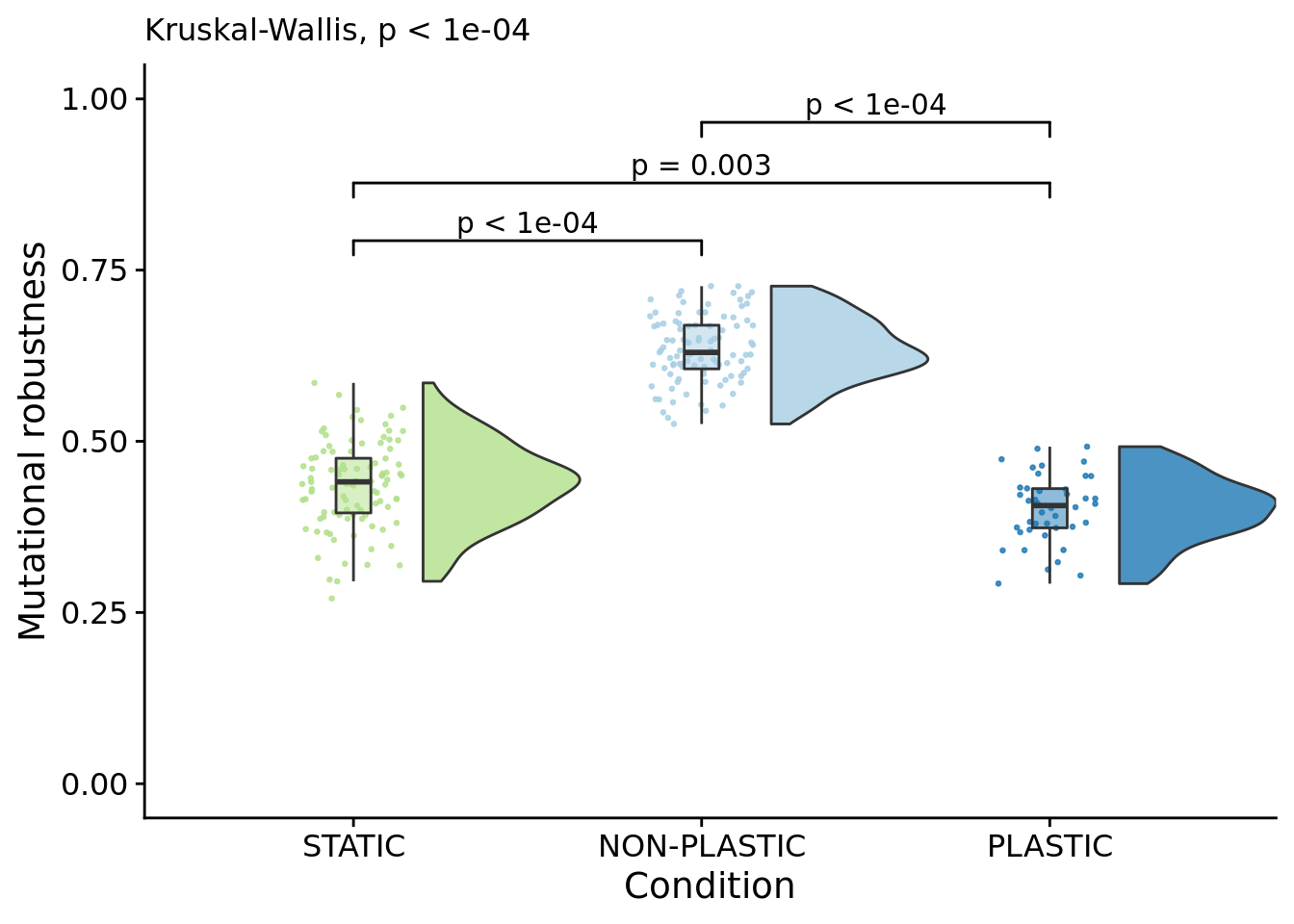
6.13 Manuscript figures
Figures styled for the paper.
magnitude_grid <- plot_grid(
coalescence_events_fig +
theme(
legend.position="none",
axis.title.x=element_blank()
) +
ggtitle("Coalescence events count"),
mutation_count_fig +
theme(
legend.position="none",
axis.title.x=element_blank()
) +
ggtitle("Mutation count"),
phenotypic_volatility_fig +
theme(
legend.position="none",
axis.title.x=element_blank()
) +
ggtitle("Phenotypic volatility"),
nrow=1,
ncol=3,
align="v",
labels="auto"
)
magnitude_grid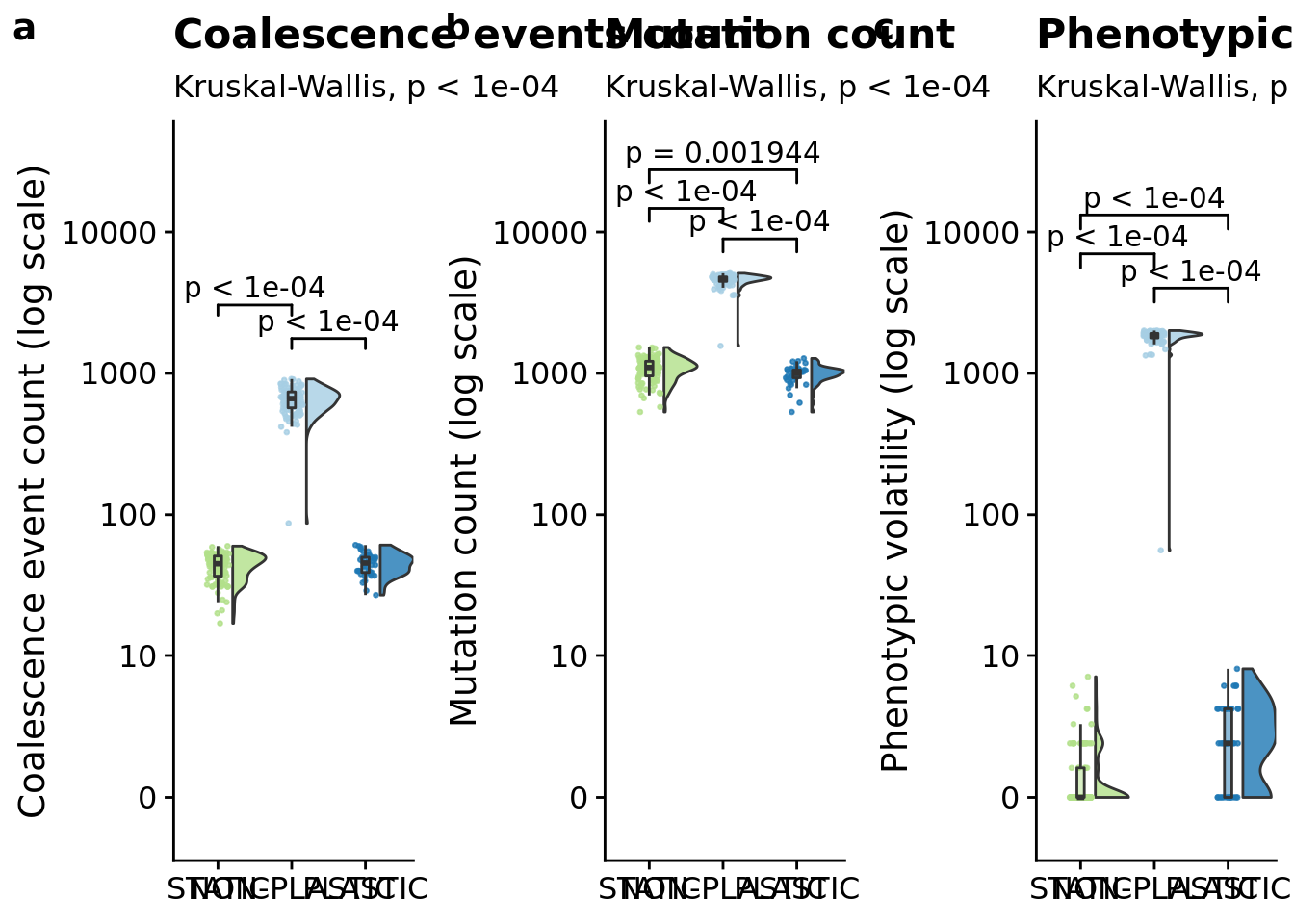
pace_grid <- plot_grid(
coalescence_events_freq_fig +
theme(
legend.position="none",
axis.title.x=element_blank()
) +
ggtitle("Generations between coalescence events"),
mutational_stability_fig +
theme(
legend.position="none",
axis.title.x=element_blank()
) +
ggtitle("Realized mutational robustness"),
nrow=1,
ncol=2,
align="v",
labels="auto"
)
pace_grid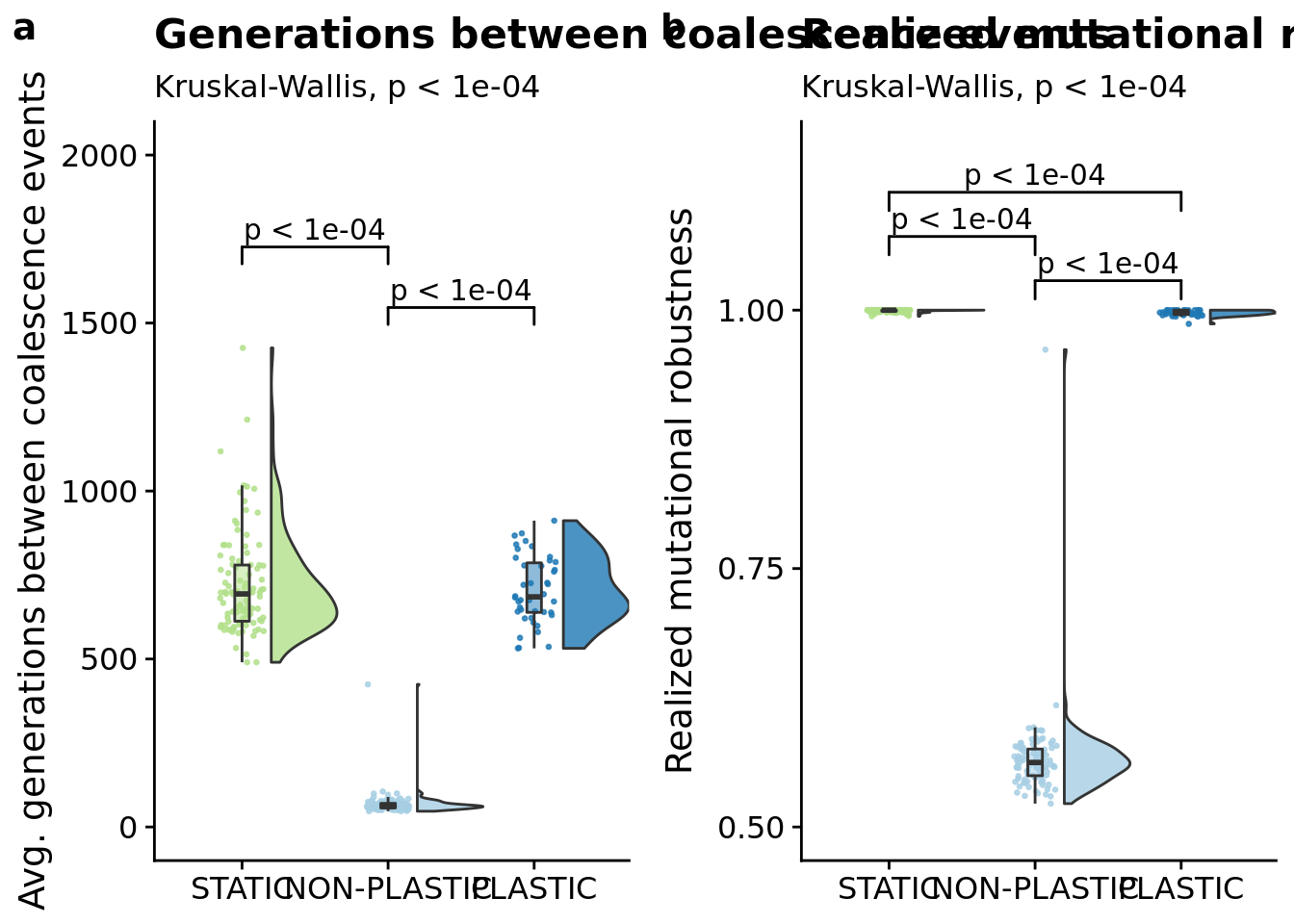
# Even though mutational robustness is shown by itself, this ensures it is plotted identically to the other multi-figure panels
mut_robustness_grid = plot_grid(
mut_robustness_fig +
theme(
legend.position="none",
axis.title.x=element_blank()
) +
ggtitle("Mutational robustness"),
nrow=1,
ncol=1,
align="v",
labels=""
)
mut_robustness_grid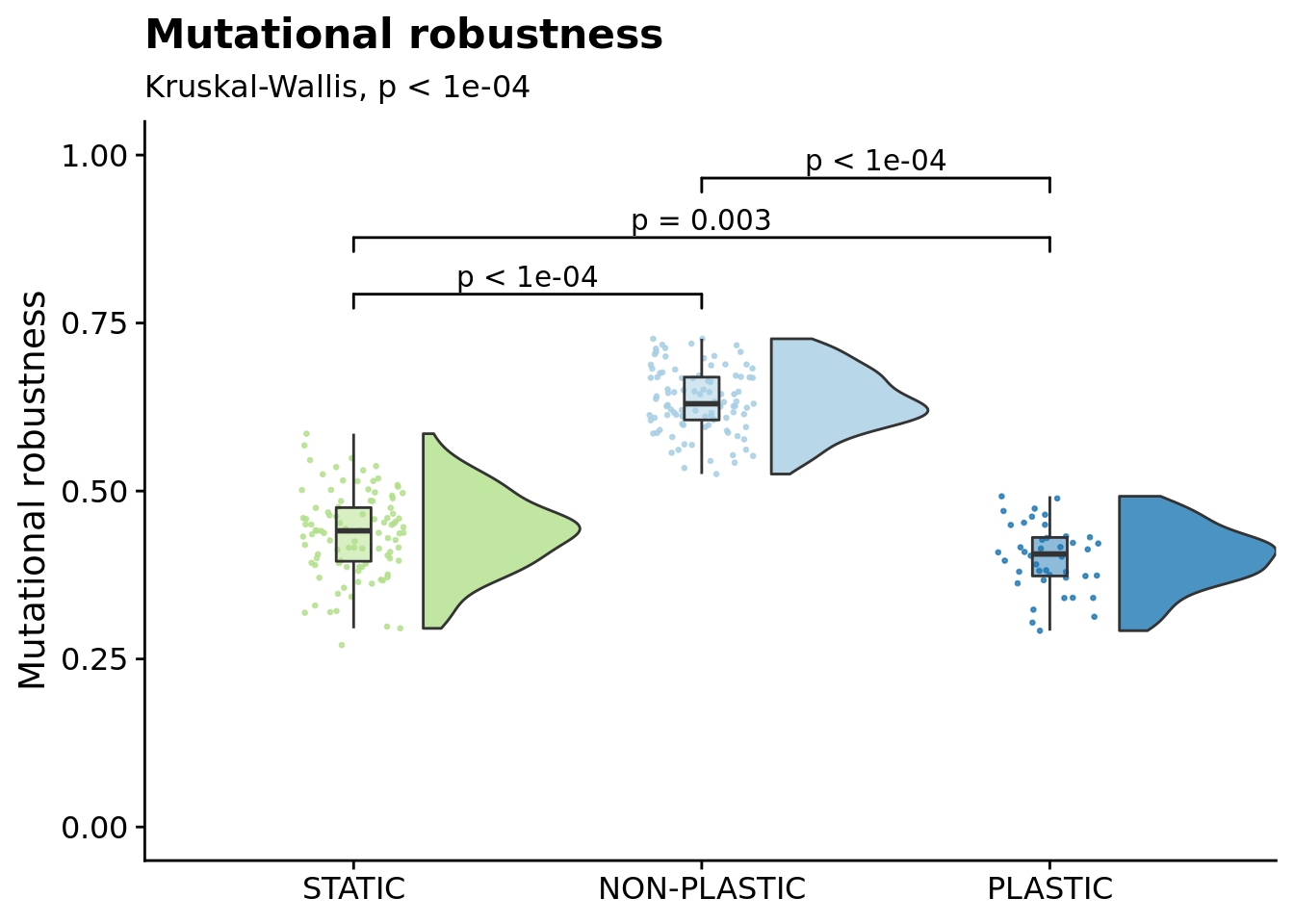
save_plot(
paste0(working_directory, "plots/", "evolutionary-change-magnitude-panel.pdf"),
magnitude_grid,
base_height=6,
base_asp=3/1
)
save_plot(
paste0(working_directory, "plots/", "evolutionary-change-pace-panel.pdf"),
pace_grid,
base_height=6,
base_asp=2/1
)
save_plot(
paste0(working_directory, "plots/", "mutational-robustness.pdf"),
mut_robustness_grid,
base_height=6,
base_asp=1
)