Chapter 10 Evolutionary change (variable length genomes)
10.1 Overview
total_updates <- 200000
replicates <- 100
all_traits <- c("not","nand","and","ornot","or","andnot")
traits_set_a <- c("not", "and", "or")
traits_set_b <- c("nand", "ornot", "andnot")
# Relative location of data.
working_directory <- "experiments/2021-01-30-evo-dynamics/analysis/" # << For bookdown
# working_directory <- "./" # << For local analysis10.2 Analysis dependencies
Load all required R libraries.
library(ggplot2)
library(tidyverse)
library(cowplot)
library(RColorBrewer)
library(Hmisc)
library(boot)
source("https://gist.githubusercontent.com/benmarwick/2a1bb0133ff568cbe28d/raw/fb53bd97121f7f9ce947837ef1a4c65a73bffb3f/geom_flat_violin.R")These analyses were conducted/knitted with the following computing environment:
## _
## platform x86_64-pc-linux-gnu
## arch x86_64
## os linux-gnu
## system x86_64, linux-gnu
## status
## major 4
## minor 1.3
## year 2022
## month 03
## day 10
## svn rev 81868
## language R
## version.string R version 4.1.3 (2022-03-10)
## nickname One Push-Up10.3 Setup
summary_data_loc <- paste0(working_directory, "data/aggregate.csv")
summary_data <- read.csv(summary_data_loc, na.strings="NONE")
summary_data$DISABLE_REACTION_SENSORS <- as.factor(summary_data$DISABLE_REACTION_SENSORS)
summary_data$chg_env <- summary_data$chg_env == "True"
summary_data$dominant_plastic_odd_even <- as.factor(summary_data$dominant_plastic_odd_even)
summary_data$sensors <- summary_data$DISABLE_REACTION_SENSORS == "0"
summary_data$is_plastic <- summary_data$dominant_plastic_odd_even == "True"
env_label_fun <- function(chg_env) {
if (chg_env) {
return("Fluctuating")
} else {
return("Constant")
}
}
sensors_label_fun <- function(has_sensors) {
if (has_sensors) {
return("Sensors")
} else {
return("No sensors")
}
}
# note that this labeler makes assumptions about how we set up our experiment
condition_label_fun <- function(has_sensors, env_chg) {
if (has_sensors && env_chg) {
return("PLASTIC")
} else if (env_chg) {
return("NON-PLASTIC")
} else {
return("STATIC")
}
}
summary_data$env_label <- mapply(
env_label_fun,
summary_data$chg_env
)
summary_data$sensors_label <- mapply(
sensors_label_fun,
summary_data$sensors
)
summary_data$condition <- mapply(
condition_label_fun,
summary_data$sensors,
summary_data$chg_env
)
condition_order = c(
"STATIC",
"NON-PLASTIC",
"PLASTIC"
)
####### misc #######
# Configure our default graphing theme
theme_set(theme_cowplot())
dir.create(paste0(working_directory, "plots"), showWarnings=FALSE)10.4 Evolution of phenotypic plasticity
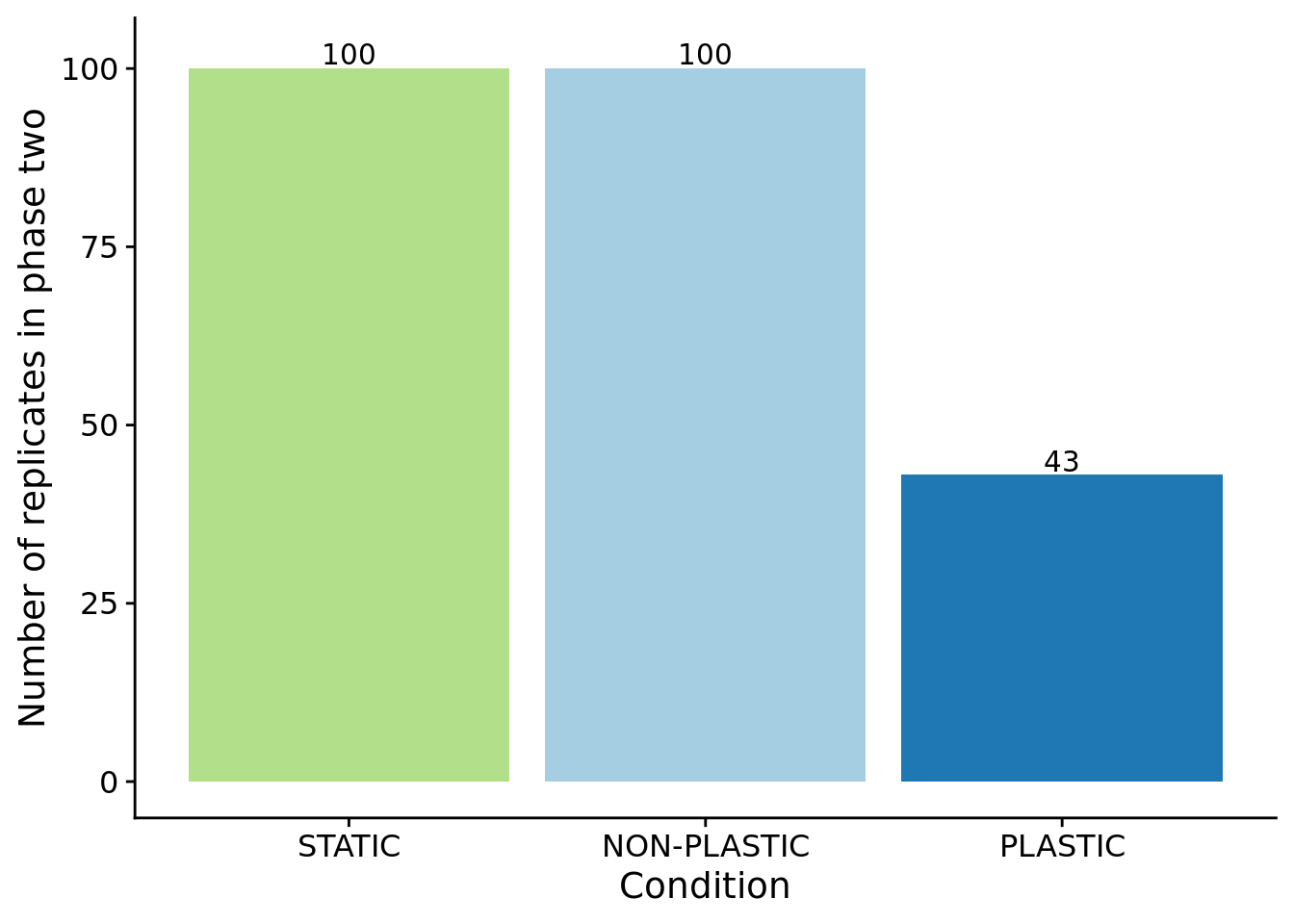
For sensor-enabled populations in fluctuating environments, we only transfered populations containing an optimally plastic genotype to phase two.
summary_data_grouped = dplyr::group_by(summary_data, sensors, env_label, condition)
summary_data_group_counts = dplyr::summarize(summary_data_grouped, n=dplyr::n())## `summarise()` has grouped output by 'sensors', 'env_label'. You can override
## using the `.groups` argument.ggplot(summary_data_group_counts, aes(x=condition, y=n, fill=condition)) +
geom_col(position=position_dodge(0.9)) +
geom_text(aes(label=n, y=n+2)) +
scale_x_discrete(
name="Condition",
limits=condition_order
) +
scale_fill_brewer(
palette="Paired"
) +
scale_color_brewer(
palette="Paired"
) +
ylab("Number of replicates in phase two") +
theme(
legend.position="none"
)
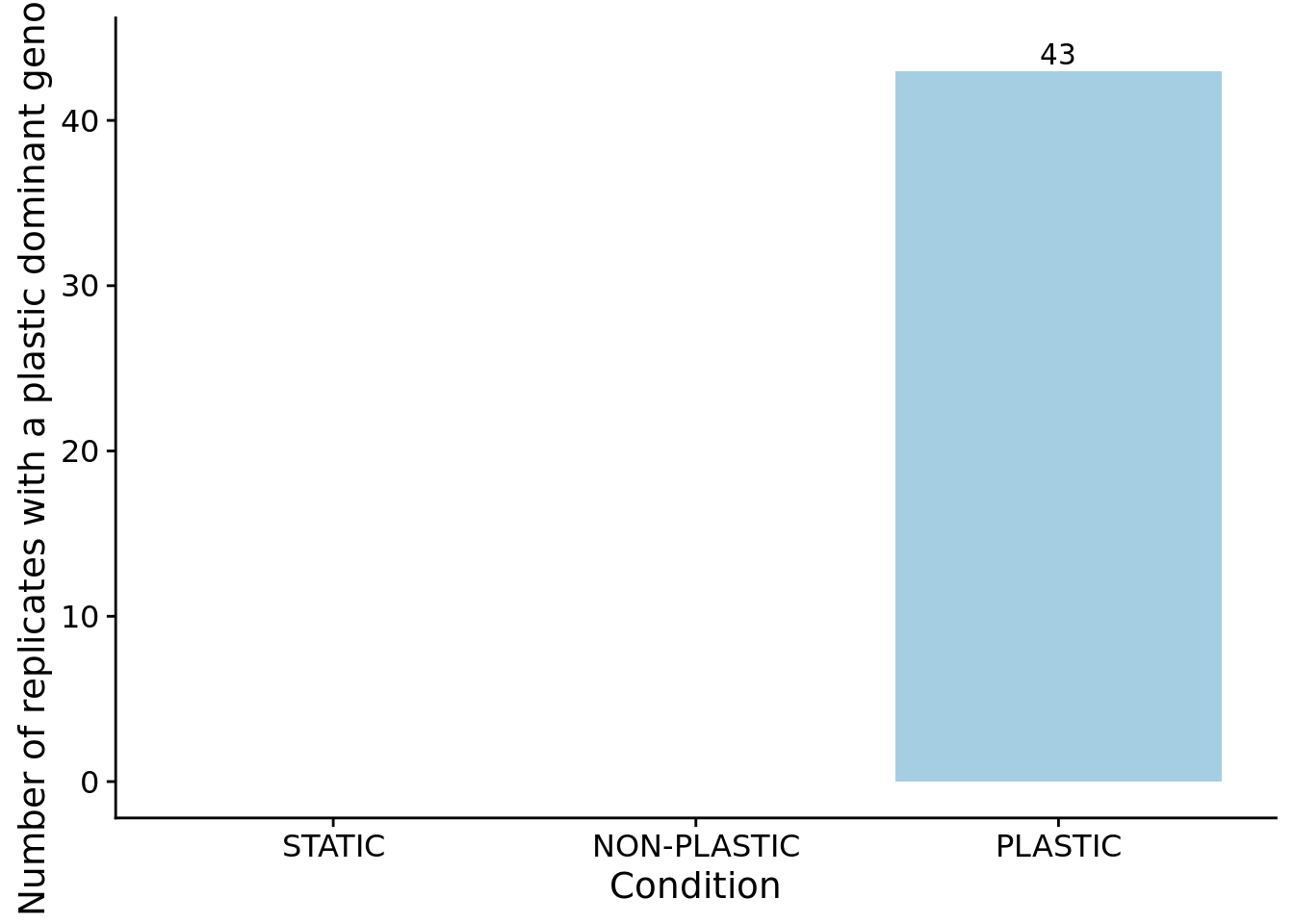
We can confirm our expectation that the dominant genotypes in non-plastic conditions are not phenotypically plastic.
summary_data_grouped = dplyr::group_by(summary_data, condition, is_plastic)
summary_data_group_counts = dplyr::summarize(summary_data_grouped, n=dplyr::n())
ggplot(filter(summary_data_group_counts, is_plastic), aes(x=condition, y=n, fill=condition)) +
geom_col(position=position_dodge(0.9)) +
scale_x_discrete(
name="Condition",
limits=condition_order
) +
scale_fill_brewer(
palette="Paired"
) +
scale_color_brewer(
palette="Paired"
) +
geom_text(aes(label=n, y=n+1)) +
ylab("Number of replicates with a plastic dominant genotype") +
theme(
legend.position="none"
)
10.5 Genome length
Single-instruction insertions and deletions were possible for this experiment, so genome size also evolved.
ggplot(summary_data, aes(x=condition, y=dominant_genome_length, fill=condition)) +
geom_flat_violin(
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Condition",
limits=condition_order
) +
scale_fill_brewer(
palette="Paired"
) +
scale_color_brewer(
palette="Paired"
) +
ylab("Genome length") +
theme(
legend.position="none"
)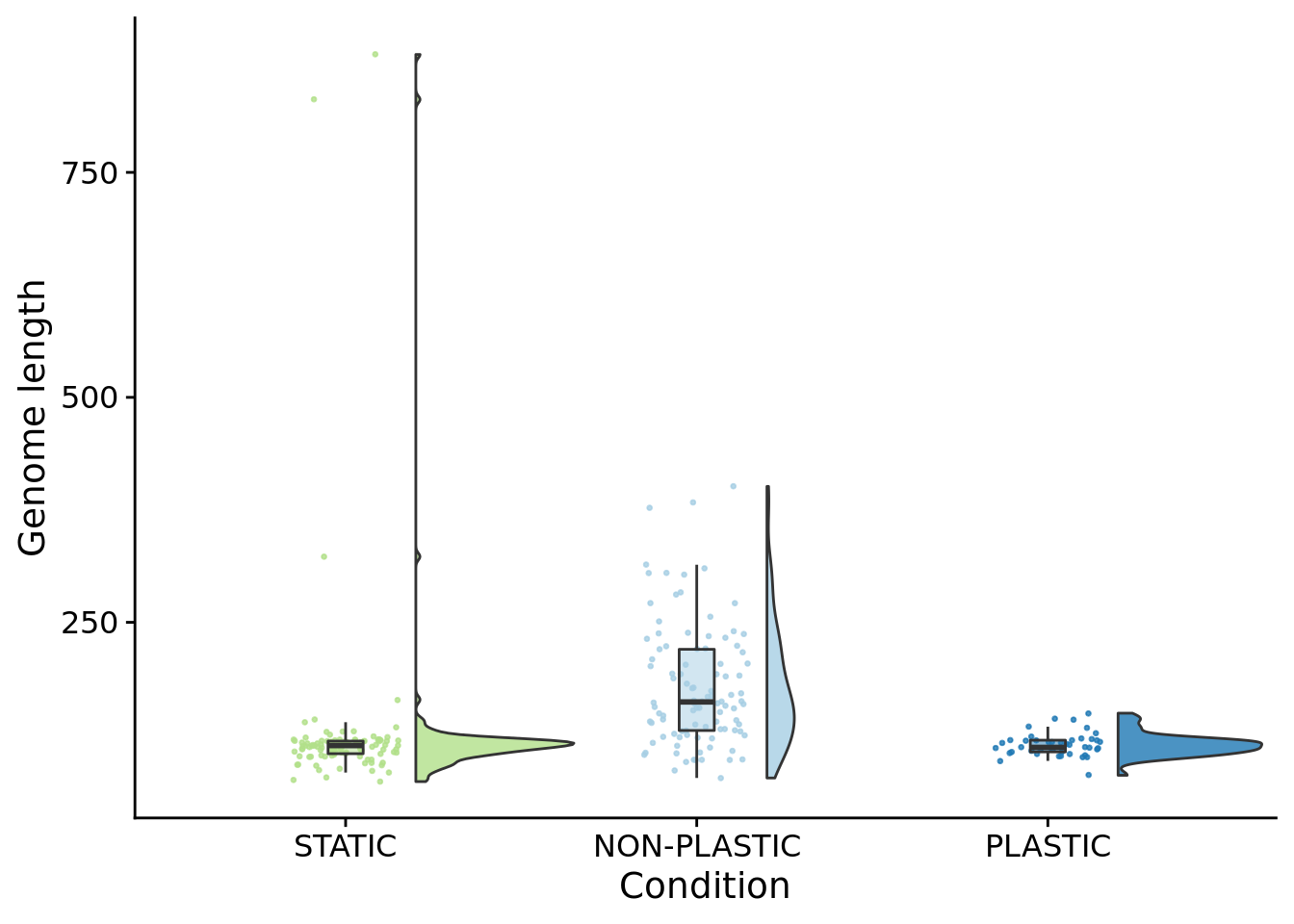
##
## Kruskal-Wallis rank sum test
##
## data: dominant_genome_length by condition
## Kruskal-Wallis chi-squared = 82.798, df = 2, p-value < 2.2e-16pairwise.wilcox.test(
x=summary_data$dominant_genome_length,
g=summary_data$condition,
p.adjust.method="bonferroni",
)##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: summary_data$dominant_genome_length and summary_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC 1.8e-10 -
## STATIC < 2e-16 1
##
## P value adjustment method: bonferroni## [1] 45## [1] 47## [1] 39310.6 Average generation
ggplot(summary_data, aes(x=condition, y=time_average_generation, fill=condition)) +
geom_flat_violin(
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Condition",
limits=condition_order
) +
scale_fill_brewer(
palette="Paired"
) +
scale_color_brewer(
palette="Paired"
) +
ylab("average generation") +
theme(
legend.position="none"
)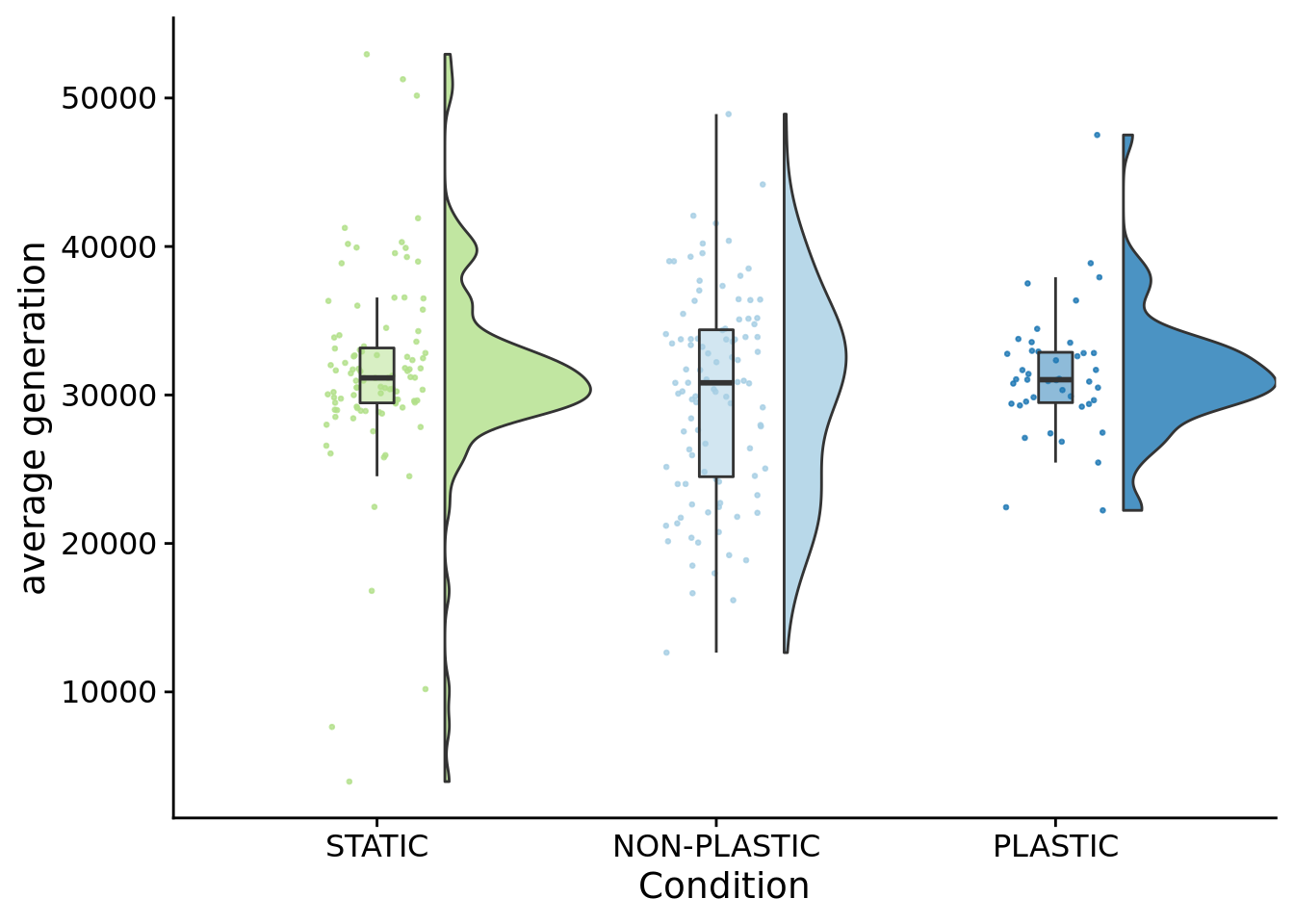
## [1] 31028.6## [1] 31147.5## [1] 30817.95##
## Kruskal-Wallis rank sum test
##
## data: time_average_generation by condition
## Kruskal-Wallis chi-squared = 1.3804, df = 2, p-value = 0.501510.7 Coalescence event count
The number of times the most recent common ancestor changes gives us the number of selective sweeps that occur during the experiment.
ggplot(summary_data, aes(x=condition, y=phylo_mrca_changes, fill=condition)) +
geom_flat_violin(
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_fill_brewer(
palette="Paired"
) +
scale_color_brewer(
palette="Paired"
) +
scale_x_discrete(
name="Condition",
limits=condition_order
) +
ylab("Coalescence event count") +
theme(
legend.position="none"
)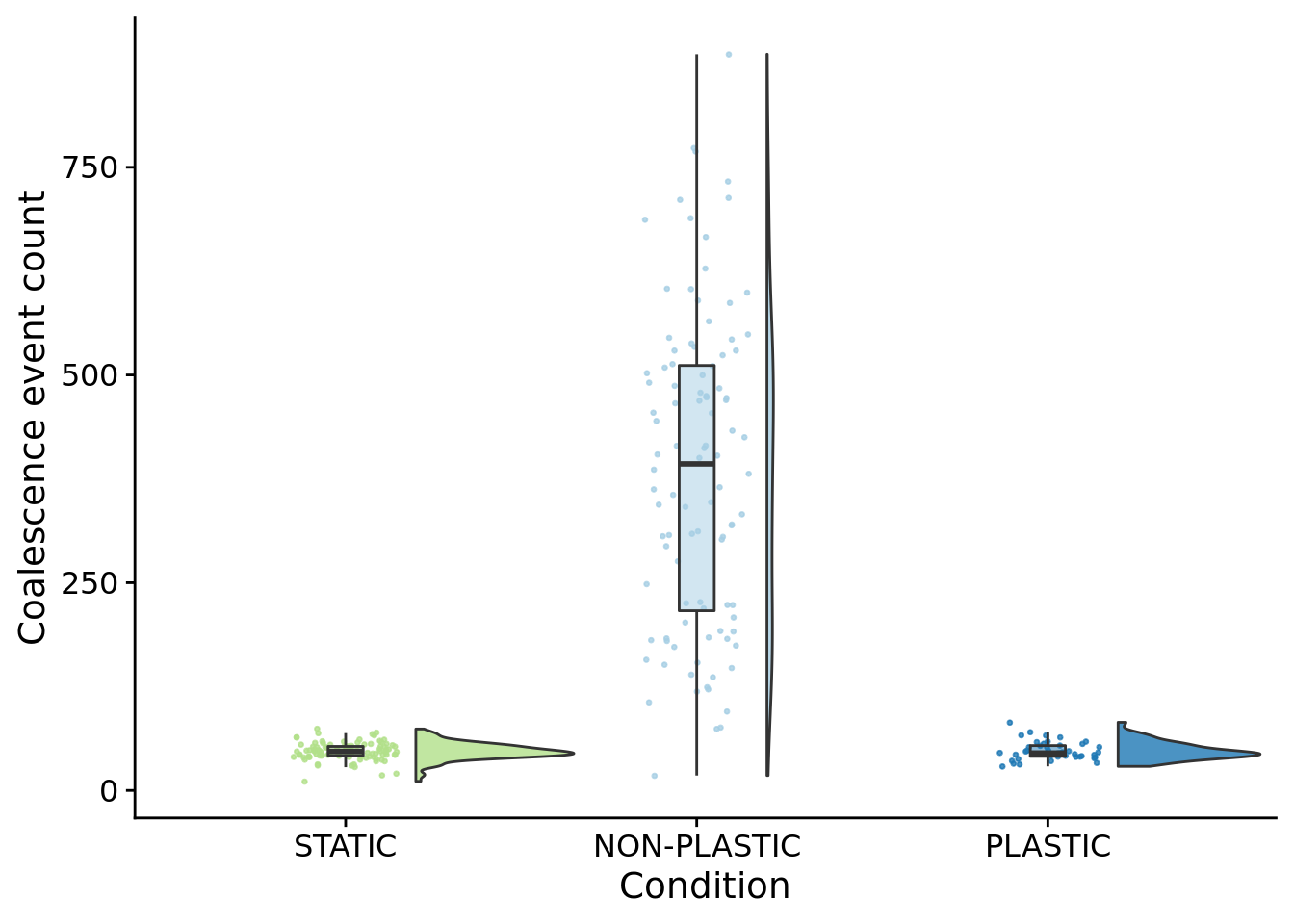
## [1] "PLASTIC: 45"## [1] "STATIC: 47"paste0(
"NON-PLASTIC: ",
median(filter(summary_data, condition=="NON-PLASTIC")$phylo_mrca_changes)
)## [1] "NON-PLASTIC: 393"##
## Kruskal-Wallis rank sum test
##
## data: phylo_mrca_changes by condition
## Kruskal-Wallis chi-squared = 168.89, df = 2, p-value < 2.2e-16pairwise.wilcox.test(
x=summary_data$phylo_mrca_changes,
g=summary_data$condition,
p.adjust.method="bonferroni",
)##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: summary_data$phylo_mrca_changes and summary_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC <2e-16 -
## STATIC <2e-16 1
##
## P value adjustment method: bonferroni10.7.1 Average number of generations between coalescence events
summary_data$generations_per_mrca_change <- summary_data$time_average_generation / summary_data$phylo_mrca_changes
ggplot(summary_data, aes(x=condition, y=generations_per_mrca_change, fill=condition)) +
geom_flat_violin(
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Condition",
limits=condition_order
) +
scale_fill_brewer(
palette="Paired"
) +
scale_color_brewer(
palette="Paired"
) +
theme(
legend.position="none"
)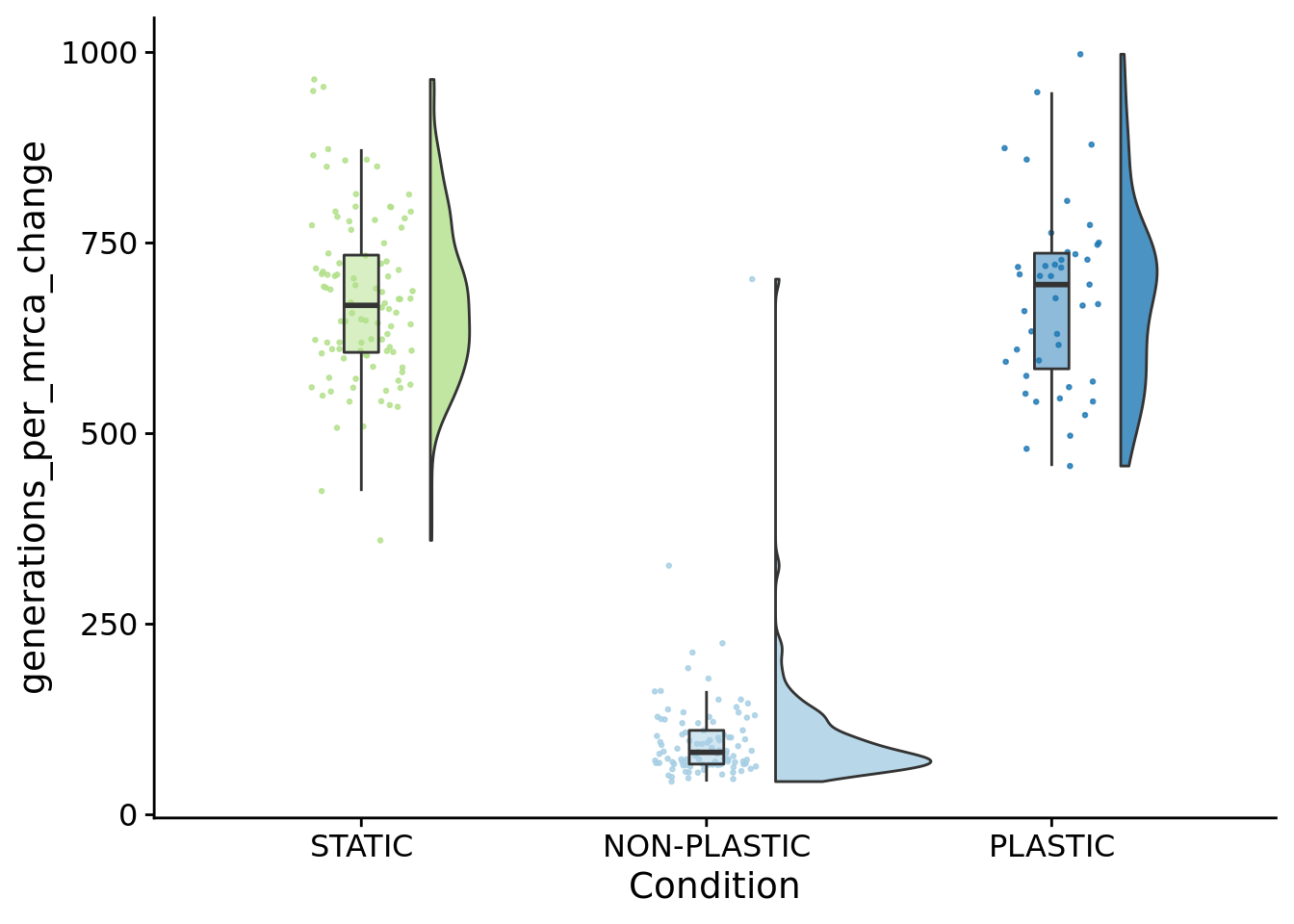
paste0(
"PLASTIC: ",
median(filter(summary_data, condition=="PLASTIC")$generations_per_mrca_change)
)## [1] "PLASTIC: 695.504761904762"## [1] "STATIC: 668.25523255814"paste0(
"NON-PLASTIC: ",
median(filter(summary_data, condition=="NON-PLASTIC")$generations_per_mrca_change)
)## [1] "NON-PLASTIC: 81.9208459944751"##
## Kruskal-Wallis rank sum test
##
## data: generations_per_mrca_change by condition
## Kruskal-Wallis chi-squared = 171.73, df = 2, p-value < 2.2e-16pairwise.wilcox.test(
x=summary_data$generations_per_mrca_change,
g=summary_data$condition,
p.adjust.method="bonferroni",
)##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: summary_data$generations_per_mrca_change and summary_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC <2e-16 -
## STATIC <2e-16 1
##
## P value adjustment method: bonferroni10.8 Phenotypic volatility along the dominant lineage
ggplot(summary_data, aes(x=condition, y=dominant_lineage_trait_volatility, fill=condition)) +
geom_flat_violin(
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Condition",
limits=condition_order
) +
scale_y_continuous(
name="Phenotypic volatility (log scale)",
trans="pseudo_log",
breaks=c(0, 10, 100, 1000, 10000),
limits=c(-1,10000)
) +
scale_fill_brewer(
palette="Paired"
) +
scale_color_brewer(
palette="Paired"
) +
theme(
legend.position="none"
)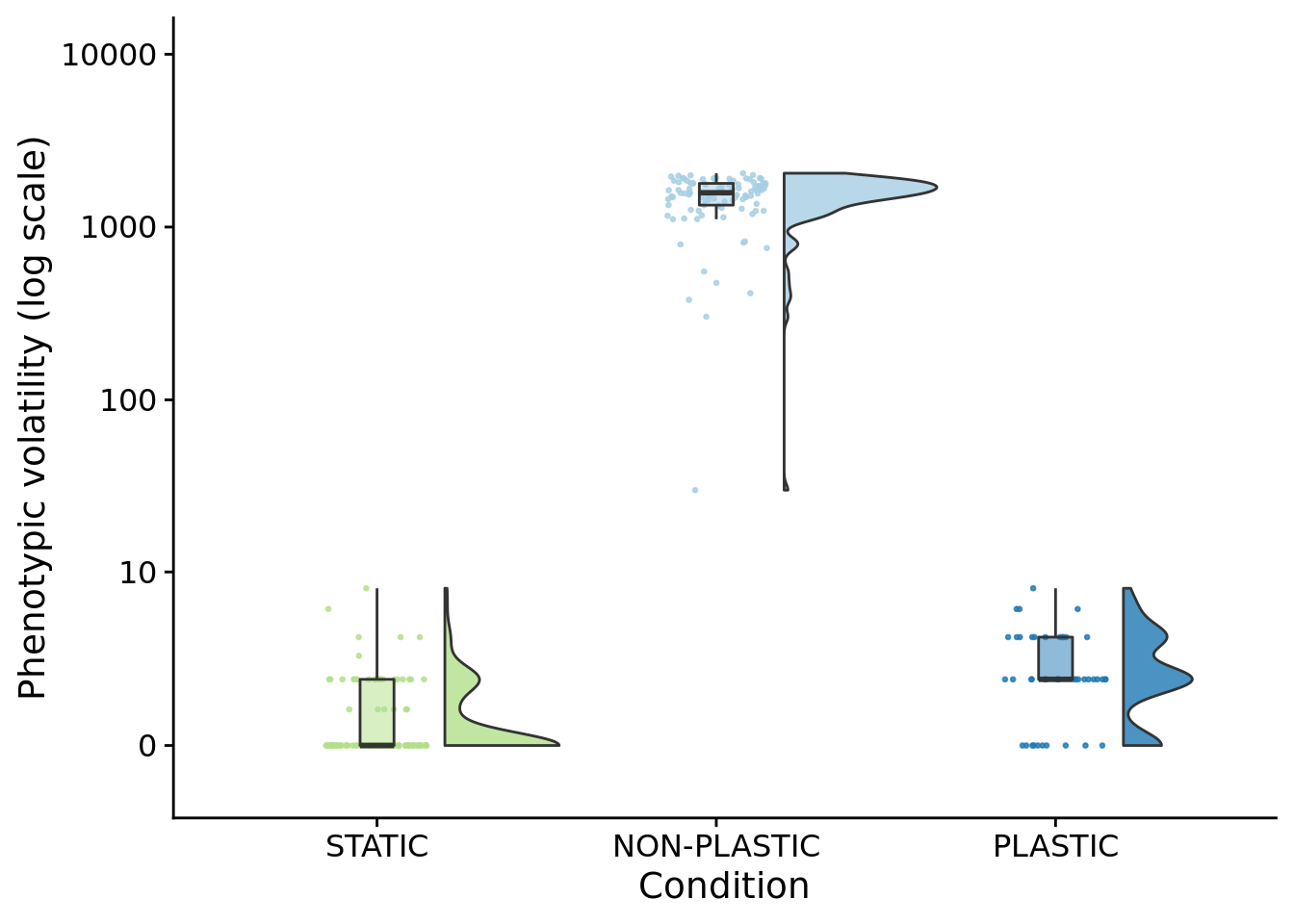
paste0(
"PLASTIC: ",
median(filter(summary_data, condition=="PLASTIC")$dominant_lineage_trait_volatility)
)## [1] "PLASTIC: 2"paste0(
"STATIC: ",
median(filter(summary_data, condition=="STATIC")$dominant_lineage_trait_volatility)
)## [1] "STATIC: 0"paste0(
"NON-PLASTIC: ",
median(filter(summary_data, condition=="NON-PLASTIC")$dominant_lineage_trait_volatility)
)## [1] "NON-PLASTIC: 1580"##
## Kruskal-Wallis rank sum test
##
## data: dominant_lineage_trait_volatility by condition
## Kruskal-Wallis chi-squared = 191.98, df = 2, p-value < 2.2e-16pairwise.wilcox.test(
x=summary_data$dominant_lineage_trait_volatility,
g=summary_data$condition,
p.adjust.method="bonferroni",
)##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: summary_data$dominant_lineage_trait_volatility and summary_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC < 2e-16 -
## STATIC < 2e-16 5.2e-08
##
## P value adjustment method: bonferroni10.9 Mutation count (along dominant lineage)
ggplot(summary_data, aes(x=condition, y=dominant_lineage_total_mut_cnt, fill=condition)) +
geom_flat_violin(
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=condition),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
ylab("Mutation accumulation") +
scale_x_discrete(
name="Condition",
limits=condition_order
) +
scale_fill_brewer(
palette="Paired"
) +
scale_color_brewer(
palette="Paired"
) +
theme(
legend.position="none"
)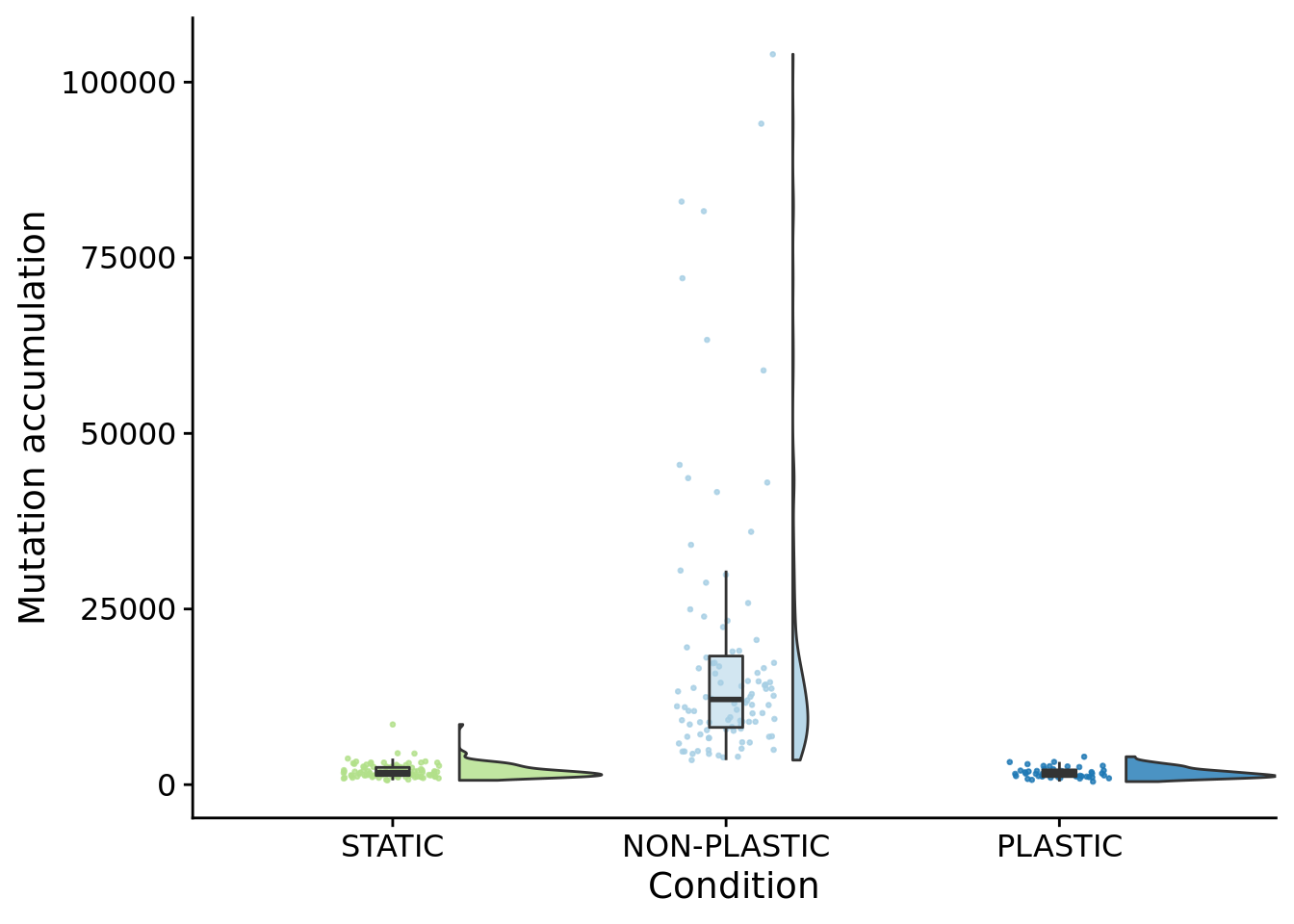
paste0(
"PLASTIC: ",
median(filter(summary_data, condition=="PLASTIC")$dominant_lineage_total_mut_cnt)
)## [1] "PLASTIC: 1552"paste0(
"STATIC: ",
median(filter(summary_data, condition=="STATIC")$dominant_lineage_total_mut_cnt)
)## [1] "STATIC: 1724.5"paste0(
"NON-PLASTIC: ",
median(filter(summary_data, condition=="NON-PLASTIC")$dominant_lineage_total_mut_cnt)
)## [1] "NON-PLASTIC: 12123"##
## Kruskal-Wallis rank sum test
##
## data: dominant_lineage_total_mut_cnt by condition
## Kruskal-Wallis chi-squared = 174.38, df = 2, p-value < 2.2e-16pairwise.wilcox.test(
x=summary_data$dominant_lineage_total_mut_cnt,
g=summary_data$condition,
p.adjust.method="bonferroni",
)##
## Pairwise comparisons using Wilcoxon rank sum test with continuity correction
##
## data: summary_data$dominant_lineage_total_mut_cnt and summary_data$condition
##
## NON-PLASTIC PLASTIC
## PLASTIC <2e-16 -
## STATIC <2e-16 0.57
##
## P value adjustment method: bonferroni