Chapter 10 Varied genome lengths
10.1 Overview
In our main experiments, we limited genome length to 100 instructions. We ran a supplemental experiment to evaluate whether this limit substantially affected the directed evolution performance. Specifically, for each of elite and lexicase selection, we ran 30 replicates of digital directed evolution with the following genome length configurations: 50, 100 (default used in our main experiments), 150, and 200.
Note that these runs were performed with a maturation period of 300 updates and run for a total of 3,000 cycles (instead of 200 and 2,000, respectively).
10.2 Analysis dependencies
Load all required R libraries
library(tidyverse)
library(ggplot2)
library(cowplot)
library(RColorBrewer)
library(scales)
library(khroma)
source("https://gist.githubusercontent.com/benmarwick/2a1bb0133ff568cbe28d/raw/fb53bd97121f7f9ce947837ef1a4c65a73bffb3f/geom_flat_violin.R")These analyses were knit with the following environment:
## _
## platform x86_64-pc-linux-gnu
## arch x86_64
## os linux-gnu
## system x86_64, linux-gnu
## status
## major 4
## minor 2.1
## year 2022
## month 06
## day 23
## svn rev 82513
## language R
## version.string R version 4.2.1 (2022-06-23)
## nickname Funny-Looking Kid10.3 Setup
Experiment summary data
exp_summary_data_loc <- paste0(working_directory,"data/experiment_summary.csv")
exp_summary_data <- read.csv(exp_summary_data_loc, na.strings="NONE")
exp_summary_data$SELECTION_METHOD <- factor(
exp_summary_data$SELECTION_METHOD,
levels=c(
"elite",
"tournament",
"lexicase",
"non-dominated-elite",
"non-dominated-tournament",
"random",
"none"
),
labels=c(
"elite",
"tourn",
"lex",
"nde",
"ndt",
"random",
"none"
)
)
exp_summary_data$AVIDAGP_ENV_FILE <- factor(
exp_summary_data$AVIDAGP_ENV_FILE,
levels=c(
"environment-no-indiv.json",
"environment-big.json"
),
labels=c(
"env-bb-0",
"env-bb-1"
)
)
exp_summary_data$NUM_POPS <- factor(
exp_summary_data$NUM_POPS,
levels=c(
"24",
"48",
"96"
)
)
exp_summary_data$UPDATES_PER_EPOCH <- as.factor(
exp_summary_data$UPDATES_PER_EPOCH
)
exp_summary_data$TOURNAMENT_SEL_TOURN_SIZE <- as.factor(
exp_summary_data$TOURNAMENT_SEL_TOURN_SIZE
)
exp_summary_data$POPULATION_SAMPLING_SIZE <- as.factor(
exp_summary_data$POPULATION_SAMPLING_SIZE
)
exp_summary_data$genome_length <- factor(
exp_summary_data$ANCESTOR_FILE,
levels=c(
"ancestor-50.gen",
"ancestor-100.gen",
"ancestor-150.gen",
"ancestor-200.gen"
),
labels=c(
"50",
"100",
"150",
"200"
)
)
exp_summary_data$SAMPLE_SIZE <- exp_summary_data$POPULATION_SAMPLING_SIZE
exp_summary_data$ENV <- exp_summary_data$AVIDAGP_ENV_FILE
exp_summary_data$U_PER_E <- exp_summary_data$UPDATES_PER_EPOCHMiscellaneous setup
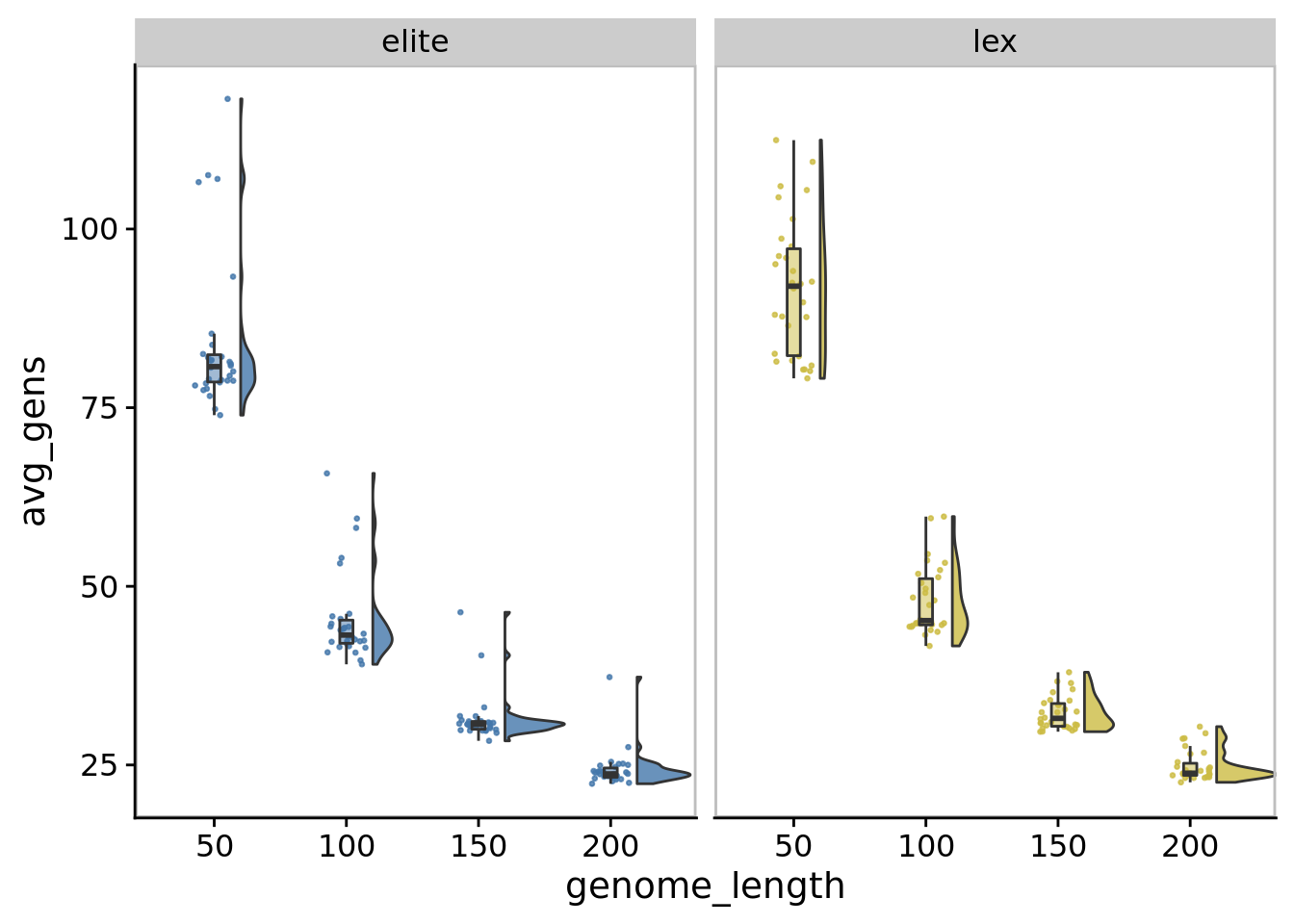
10.4 Average generations per maturation period
ggplot(
exp_summary_data,
aes(
x=genome_length,
y=avg_gens,
fill=SELECTION_METHOD
)
) +
geom_flat_violin(
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=SELECTION_METHOD),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_fill_manual(
values=selection_methods_smaller_set_colors
) +
scale_color_manual(
values=selection_methods_smaller_set_colors
) +
facet_wrap(
~SELECTION_METHOD
) +
theme(
legend.position="none",
panel.border=element_rect(colour="grey",size=1)
)
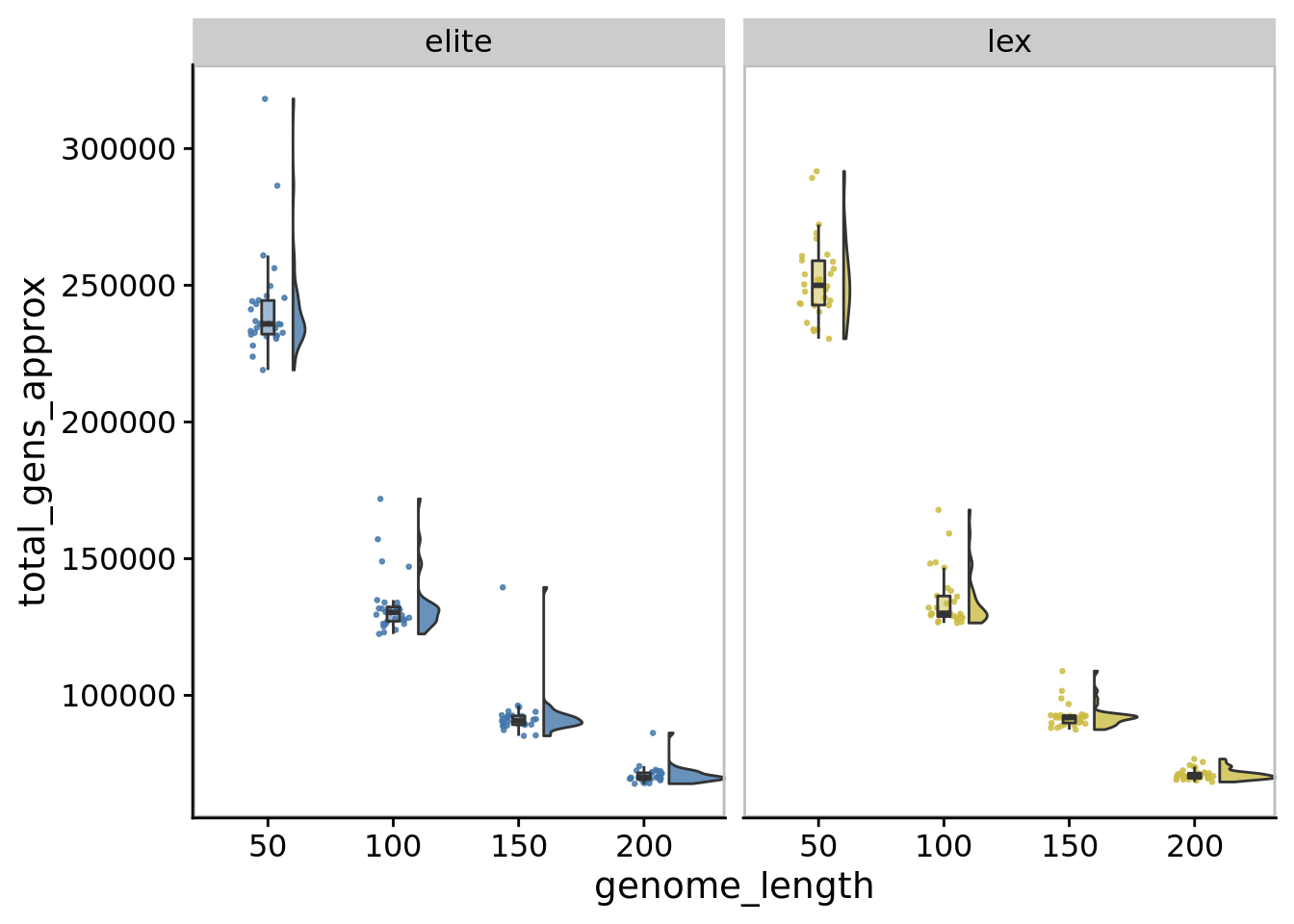
10.5 Total generations over experiment
ggplot(
exp_summary_data,
aes(
x=genome_length,
y=total_gens_approx,
fill=SELECTION_METHOD
)
) +
geom_flat_violin(
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=SELECTION_METHOD),
position = position_jitter(width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_fill_manual(
values=selection_methods_smaller_set_colors
) +
scale_color_manual(
values=selection_methods_smaller_set_colors
) +
facet_wrap(
~SELECTION_METHOD
) +
theme(
legend.position="none",
panel.border=element_rect(colour="grey",size=1)
)
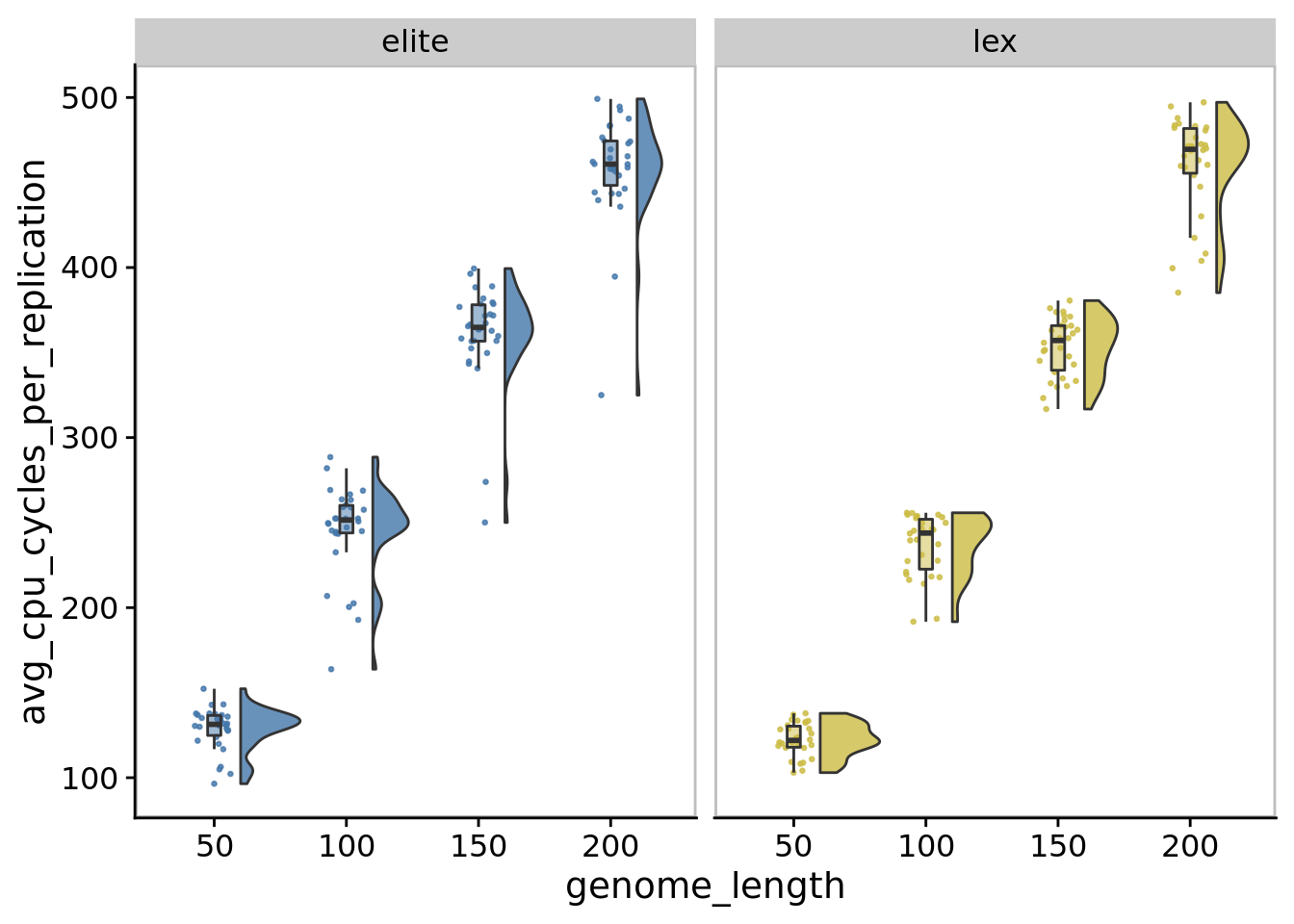
10.6 Performance
10.6.1 CPU cycles per replication
ggplot(
exp_summary_data,
aes(
x=genome_length,
y=avg_cpu_cycles_per_replication,
fill=SELECTION_METHOD
)
) +
geom_flat_violin(
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=SELECTION_METHOD),
position = position_jitter(height=0, width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_fill_manual(
values=selection_methods_smaller_set_colors
) +
scale_color_manual(
values=selection_methods_smaller_set_colors
) +
facet_wrap(
~SELECTION_METHOD
) +
theme(
legend.position="none",
panel.border=element_rect(colour="grey",size=1)
)
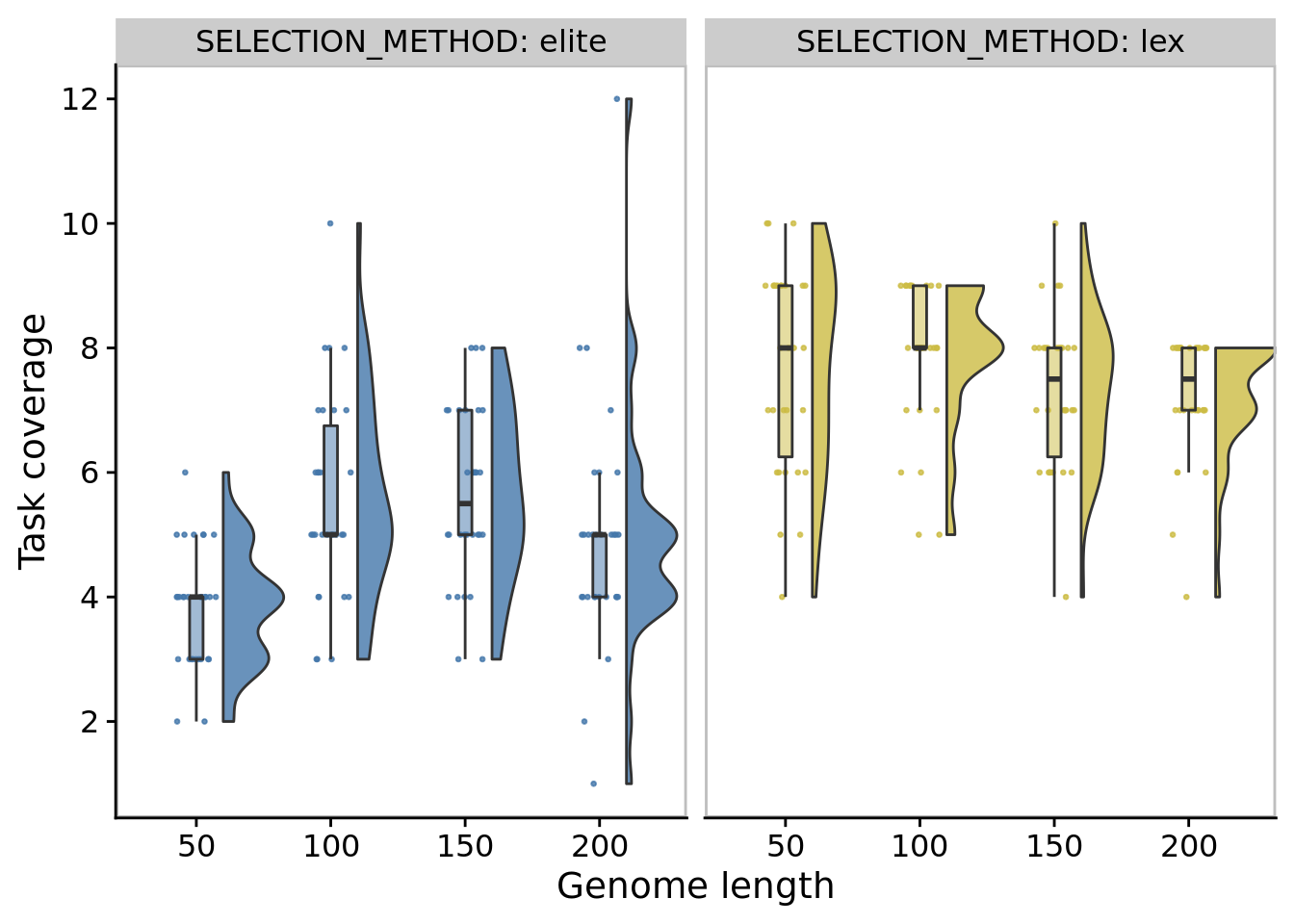
## Saving 7 x 5 in image10.6.2 Best single-population task coverage
ggplot(
exp_summary_data,
aes(
x=genome_length,
y=max_trait_coverage,
fill=SELECTION_METHOD
)
) +
geom_flat_violin(
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=SELECTION_METHOD),
position = position_jitter(height=0, width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Genome length"
) +
scale_y_continuous(
name="Task coverage",
breaks=seq(0,18,2)
) +
scale_fill_manual(
values=selection_methods_smaller_set_colors
) +
scale_color_manual(
values=selection_methods_smaller_set_colors
) +
facet_wrap(
~SELECTION_METHOD,
nrow=1,
labeller=label_both
) +
theme(
legend.position="none",
panel.border=element_rect(colour="grey",size=1)
)
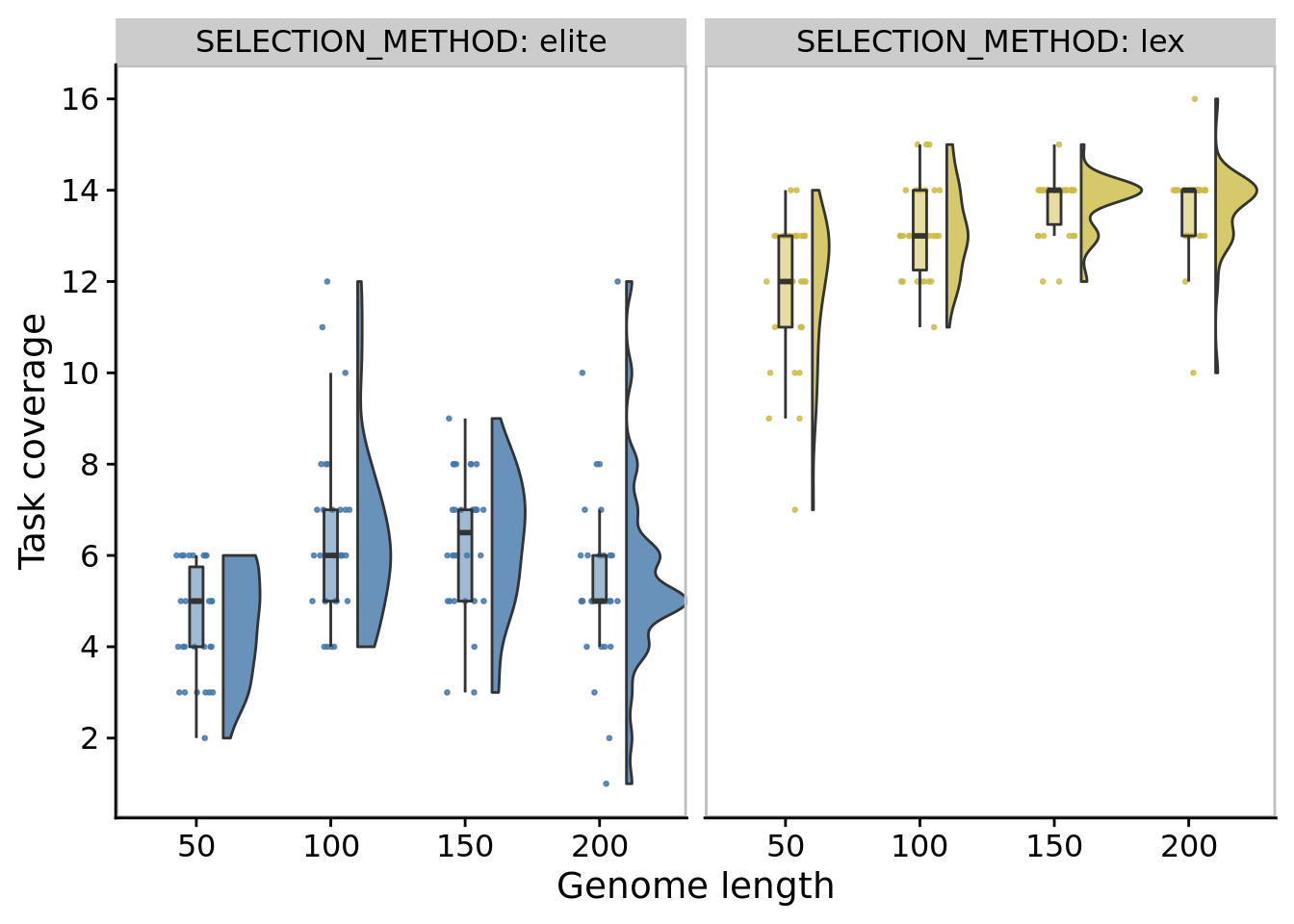
10.6.3 Metapopulation task coverage
ggplot(
exp_summary_data,
aes(
x=genome_length,
y=total_trait_coverage,
fill=SELECTION_METHOD
)
) +
geom_flat_violin(
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=SELECTION_METHOD),
position = position_jitter(height=0, width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_x_discrete(
name="Genome length"
) +
scale_y_continuous(
name="Task coverage",
breaks=seq(0,18,2)
) +
scale_fill_manual(
values=selection_methods_smaller_set_colors
) +
scale_color_manual(
values=selection_methods_smaller_set_colors
) +
facet_wrap(
~SELECTION_METHOD,
nrow=1,
labeller=label_both
) +
theme(
legend.position="none",
panel.border=element_rect(colour="grey",size=1)
)
10.7 Population-level task profile diversity
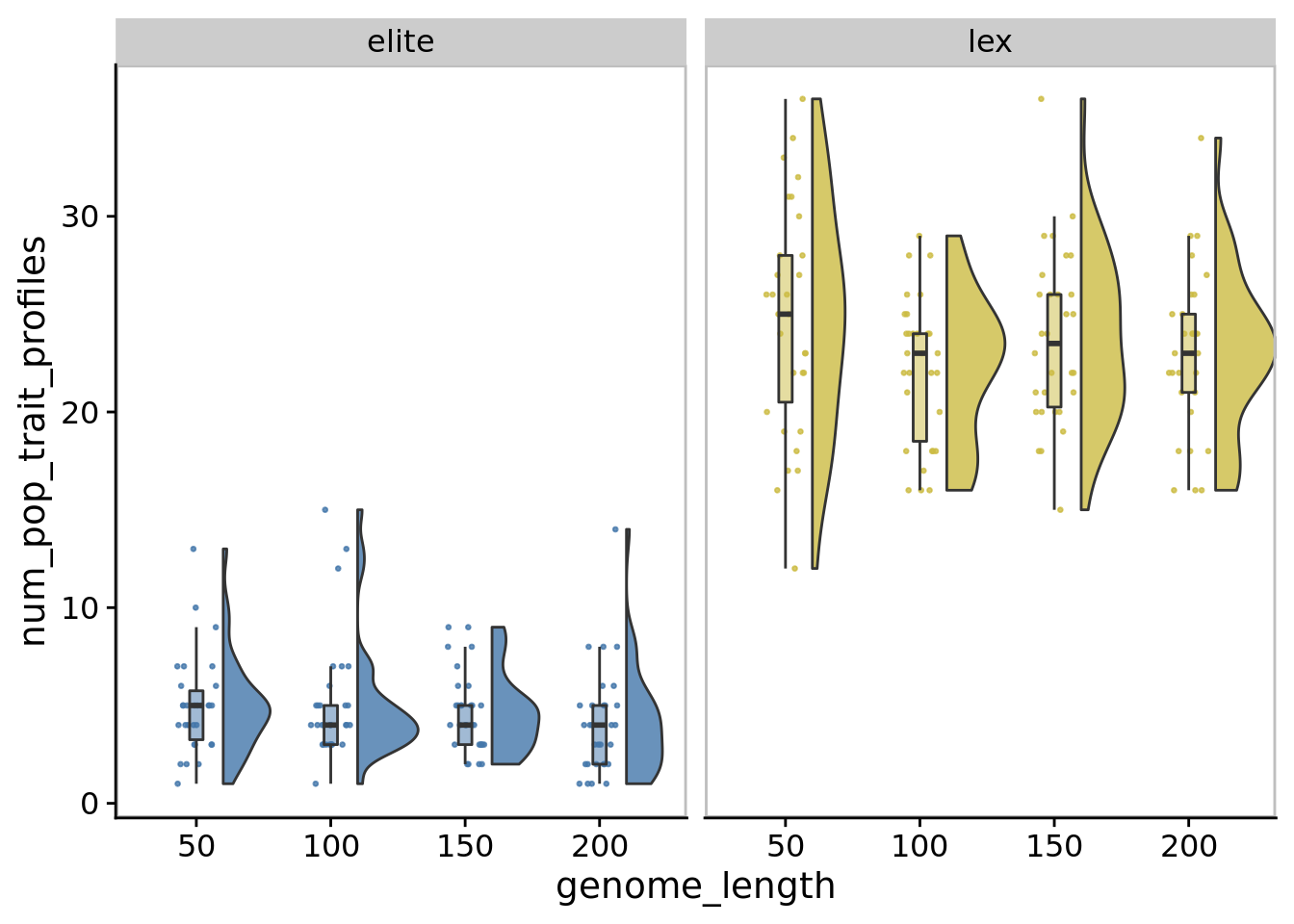
10.7.1 Task profile richness
ggplot(
exp_summary_data,
aes(
x=genome_length,
y=num_pop_trait_profiles,
fill=SELECTION_METHOD
)
) +
geom_flat_violin(
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=SELECTION_METHOD),
position = position_jitter(height=0, width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_fill_manual(
values=selection_methods_smaller_set_colors
) +
scale_color_manual(
values=selection_methods_smaller_set_colors
) +
facet_wrap(
~SELECTION_METHOD
) +
theme(
legend.position="none",
panel.border=element_rect(colour="grey",size=1)
)
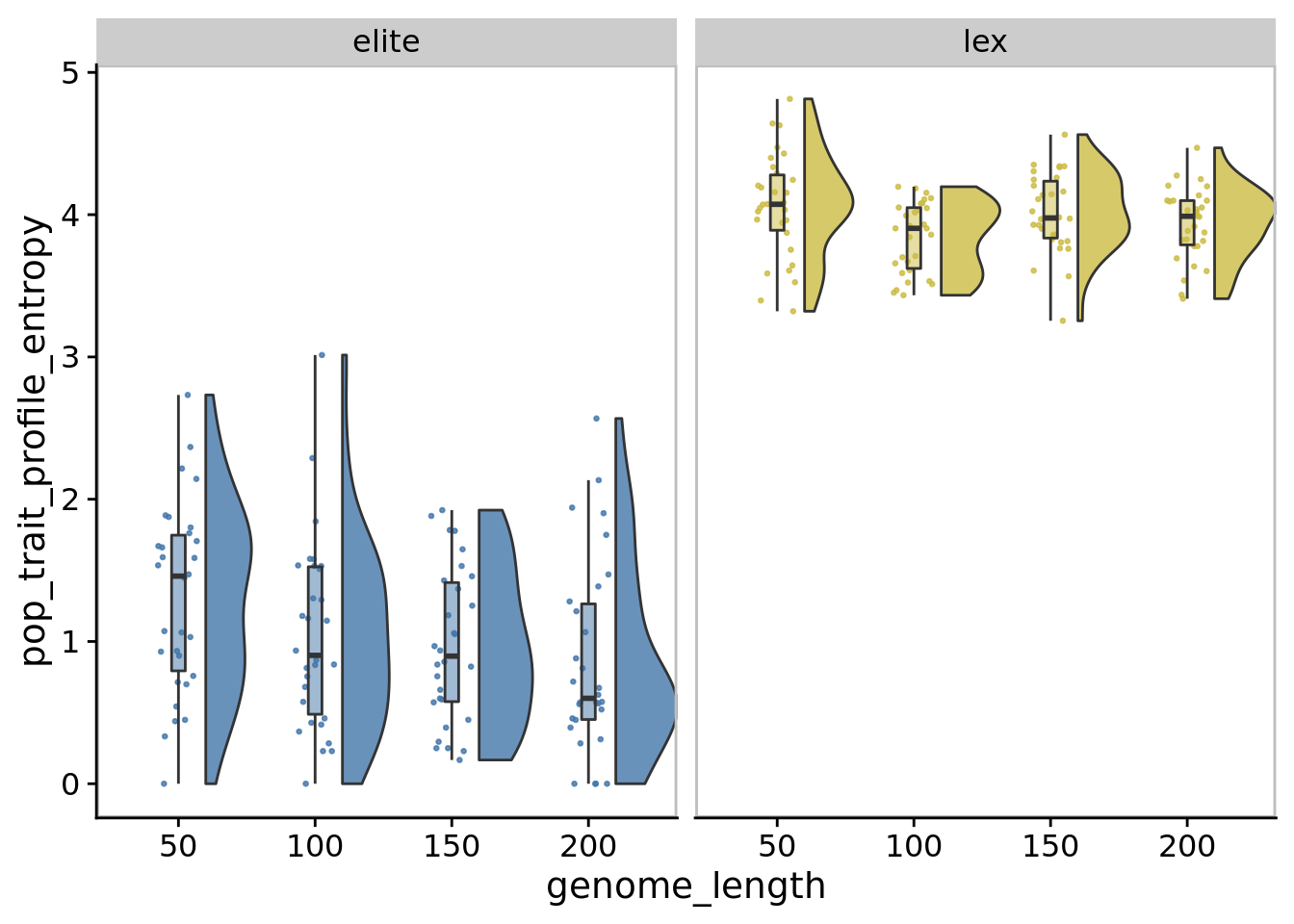
## Saving 7 x 5 in image10.7.2 Task profile entropy
ggplot(
exp_summary_data,
aes(
x=genome_length,
y=pop_trait_profile_entropy,
fill=SELECTION_METHOD
)
) +
geom_flat_violin(
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=SELECTION_METHOD),
position = position_jitter(height=0, width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_fill_manual(
values=selection_methods_smaller_set_colors
) +
scale_color_manual(
values=selection_methods_smaller_set_colors
) +
facet_wrap(
~SELECTION_METHOD
) +
theme(
legend.position="none",
panel.border=element_rect(colour="grey",size=1)
)
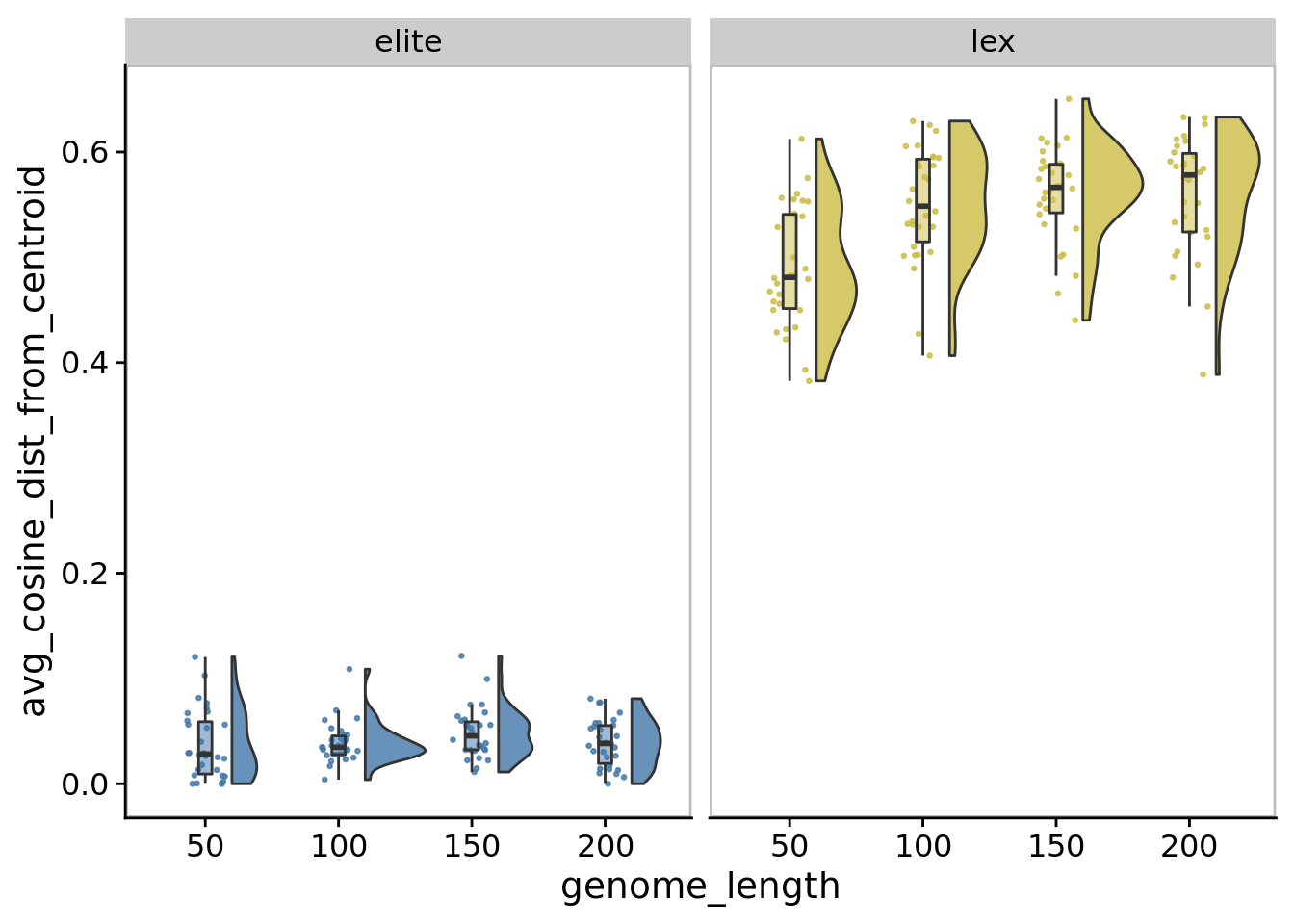
## Saving 7 x 5 in image10.7.3 Spread (avg cosine distance)
ggplot(
exp_summary_data,
aes(
x=genome_length,
y=avg_cosine_dist_from_centroid,
fill=SELECTION_METHOD
)
) +
geom_flat_violin(
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=SELECTION_METHOD),
position = position_jitter(height=0, width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_fill_manual(
values=selection_methods_smaller_set_colors
) +
scale_color_manual(
values=selection_methods_smaller_set_colors
) +
facet_wrap(
~SELECTION_METHOD
) +
theme(
legend.position="none",
panel.border=element_rect(colour="grey",size=1)
)
## Saving 7 x 5 in image10.8 Selection
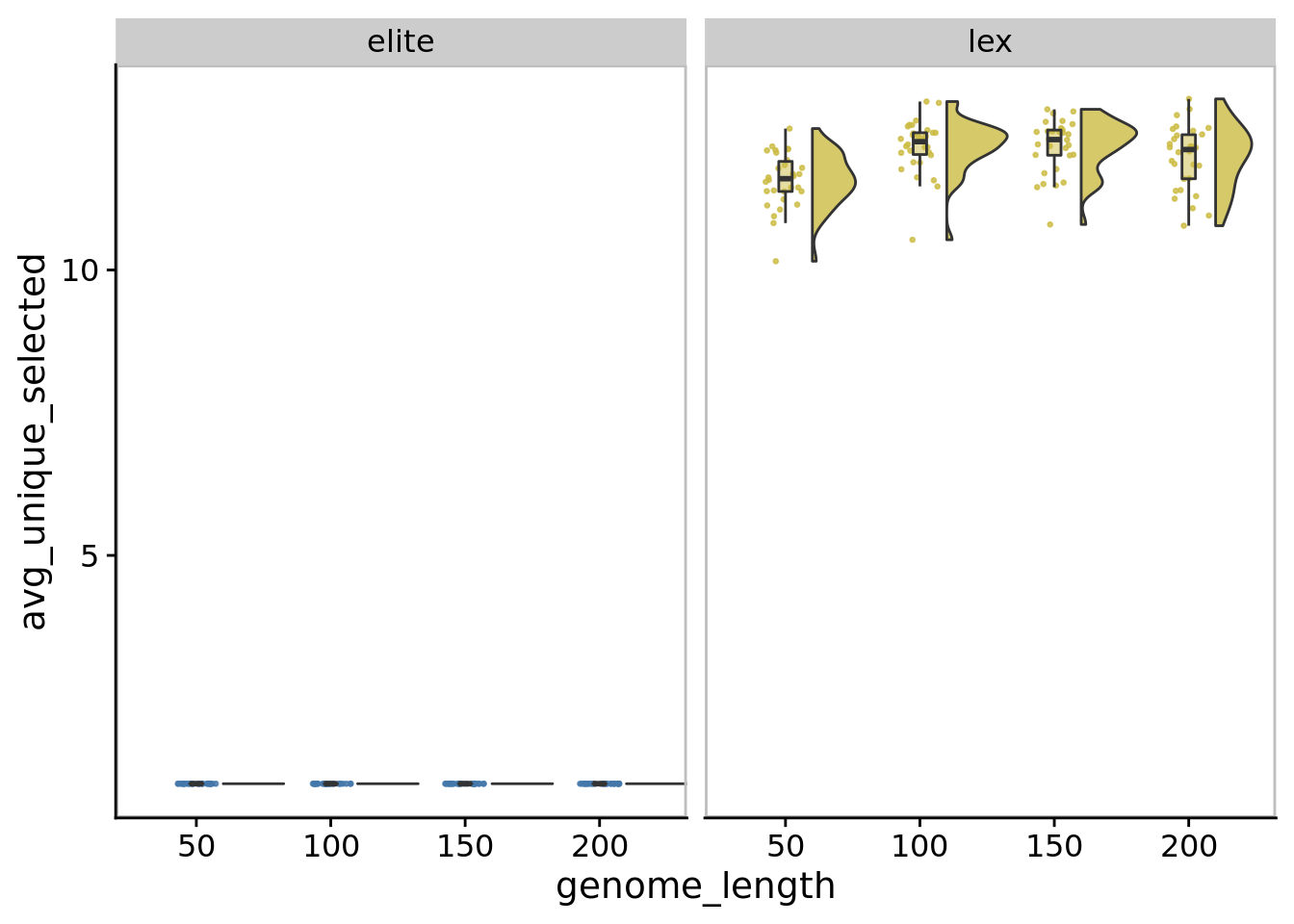
10.8.1 Average number of unique populations selected
ggplot(
exp_summary_data,
aes(
x=genome_length,
y=avg_unique_selected,
fill=SELECTION_METHOD
)
) +
geom_flat_violin(
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=SELECTION_METHOD),
position = position_jitter(height=0, width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_fill_manual(
values=selection_methods_smaller_set_colors
) +
scale_color_manual(
values=selection_methods_smaller_set_colors
) +
facet_wrap(
~SELECTION_METHOD
) +
theme(
legend.position="none",
panel.border=element_rect(colour="grey",size=1)
)
## Saving 7 x 5 in image10.8.2 Average entropy of selection ids
ggplot(
exp_summary_data,
aes(
x=genome_length,
y=avg_entropy_selected,
fill=SELECTION_METHOD
)
) +
geom_flat_violin(
position = position_nudge(x = .2, y = 0),
alpha = .8
) +
geom_point(
mapping=aes(color=SELECTION_METHOD),
position = position_jitter(height=0, width = .15),
size = .5,
alpha = 0.8
) +
geom_boxplot(
width = .1,
outlier.shape = NA,
alpha = 0.5
) +
scale_fill_manual(
values=selection_methods_smaller_set_colors
) +
scale_color_manual(
values=selection_methods_smaller_set_colors
) +
facet_wrap(
~SELECTION_METHOD
) +
theme(
legend.position="none",
panel.border=element_rect(colour="grey",size=1)
)
## Saving 7 x 5 in image